Splicing modulation by compounds with antitumour properties
Targeted modulation of alternative splicing patterns using small molecule compounds with cytostatic properties has emerged as a promising route for cancer therapy. We recently described the crystal structures of SF3B in complex with Pladienolide B - an antitumour compound representative for a broader class of modulators.
The structures explain how modulators act as competitive branch-site antagonists and provide insight into the correlation between specific chemical features of the compounds and their distinctive effects in splicing. Overall, description of their molecular recognition and mechanism of action paves the way towards the structure-based design of next-generation splicing modulators (Cretu et al., Mol. Cell, 2018).
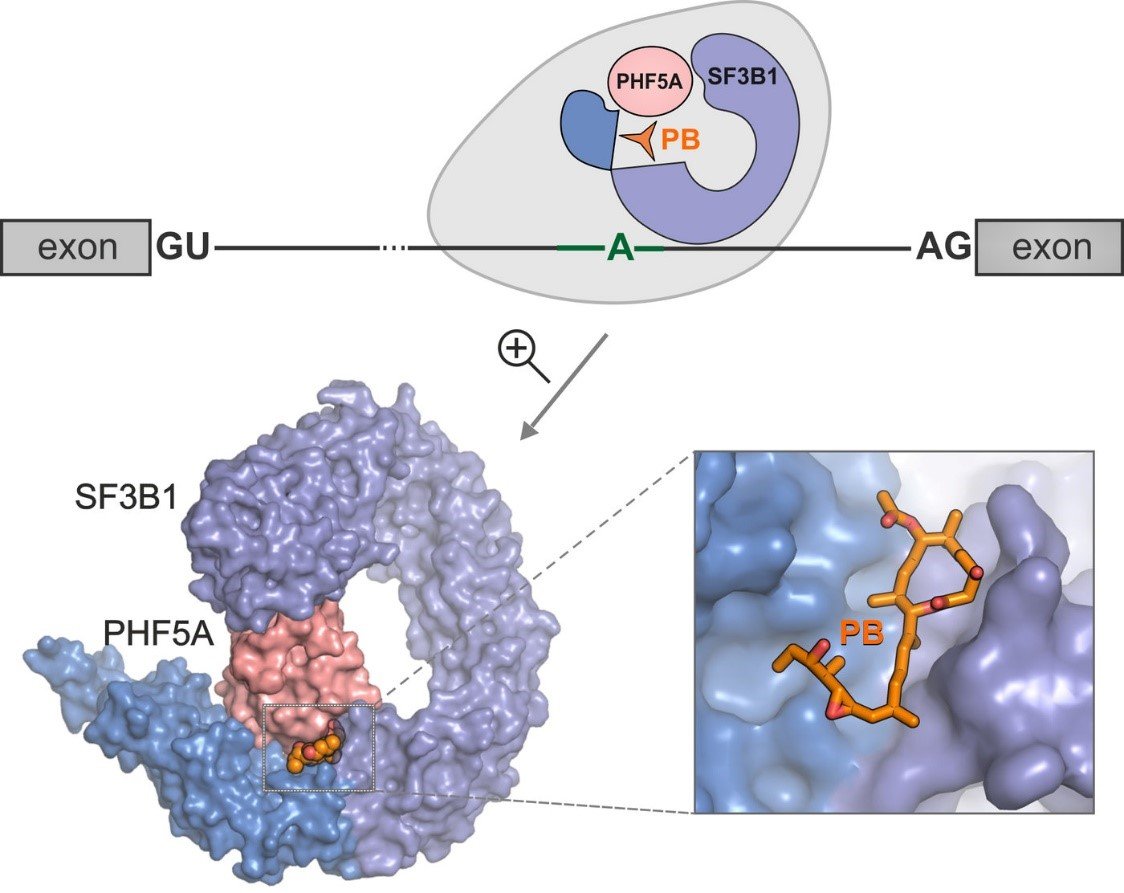
Pladienolide B binds SF3B, preventing formation of the BS-binding pocket and the transition to the closed conformation.
NTC promotes ubiquitination in splicing and DNA repair
Prp19 is a homotetramer complex involved in very diverse cellular processes, such as splicing, DNA repair, DNA transcription, protein degradation and biogenesis of lipid droplets. As part of the heteromeric Nineteen Complex (NTC), Prp19 promotes ubiquitination during formation of the spliceosomal tri-snRNP particle, and during the DNA damage response.
We showed by X-ray crystallography and functional analyses in vitro and in vivo that Prp19 is an autoinhibited E3 ubiquitin ligase that becomes active only in stable association with three other NTC components that exert specific effects on Prp19's conformation. (De Moura et al., Mol. Cell, 2018).
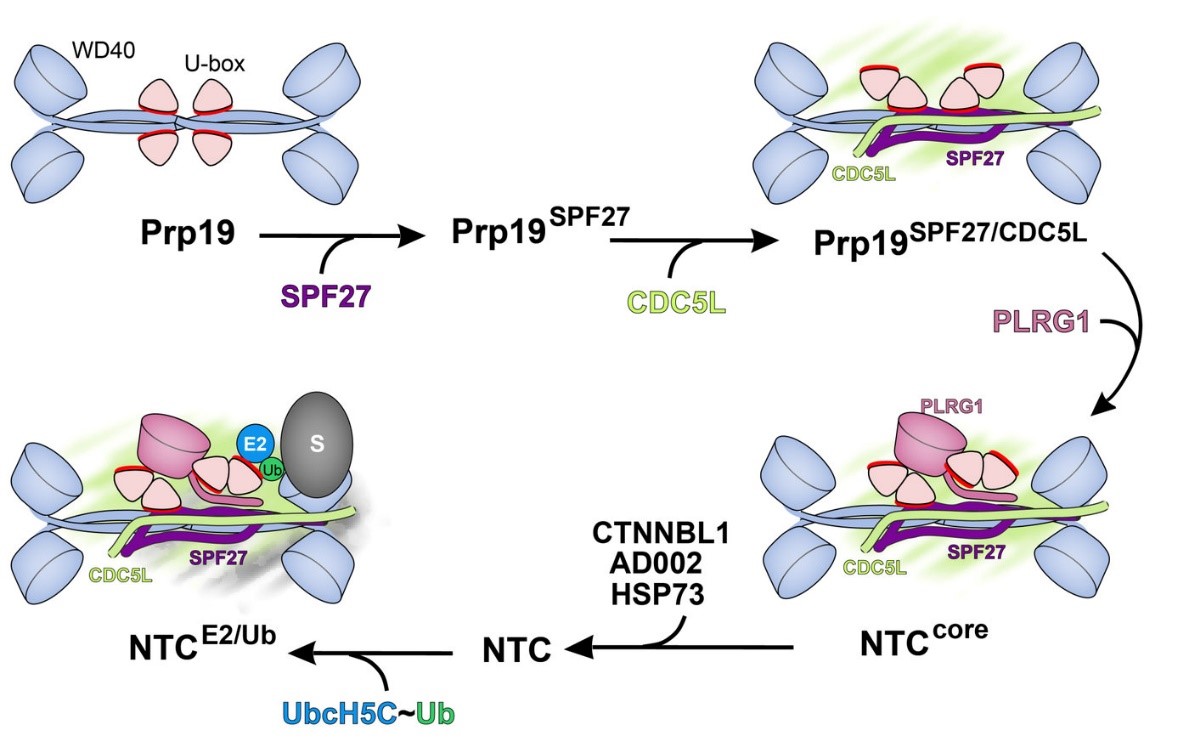
Prp19 is an autoinhibited E3 ligase activated by step-wise assembly of three splicing factors.
Helicases: the main driving forces of the splicing cycle
Eight helicases, which are conserved from yeast to humans, are sufficient for driving splicing in S. cerevisiae. However, we found that the human helicase Aquarius induces an additional ATP-dependent remodeling of the spliceosome, implying a more complex splicing pathway in humans than in yeast.
We determined the structure of Aquarius and showed that this helicase is recruited to the spliceosome as part of the pentameric intron binding complex (IBC). Aquarius is also known to act as a molecular linker that couples the splicing of pre‑mRNA to the formation of box C/D small nucleolar ribonucleoproteins (snoRNPs) and to the deposition of the exon junction complex (EJC). Our research focuses on the structural basis of these connections.
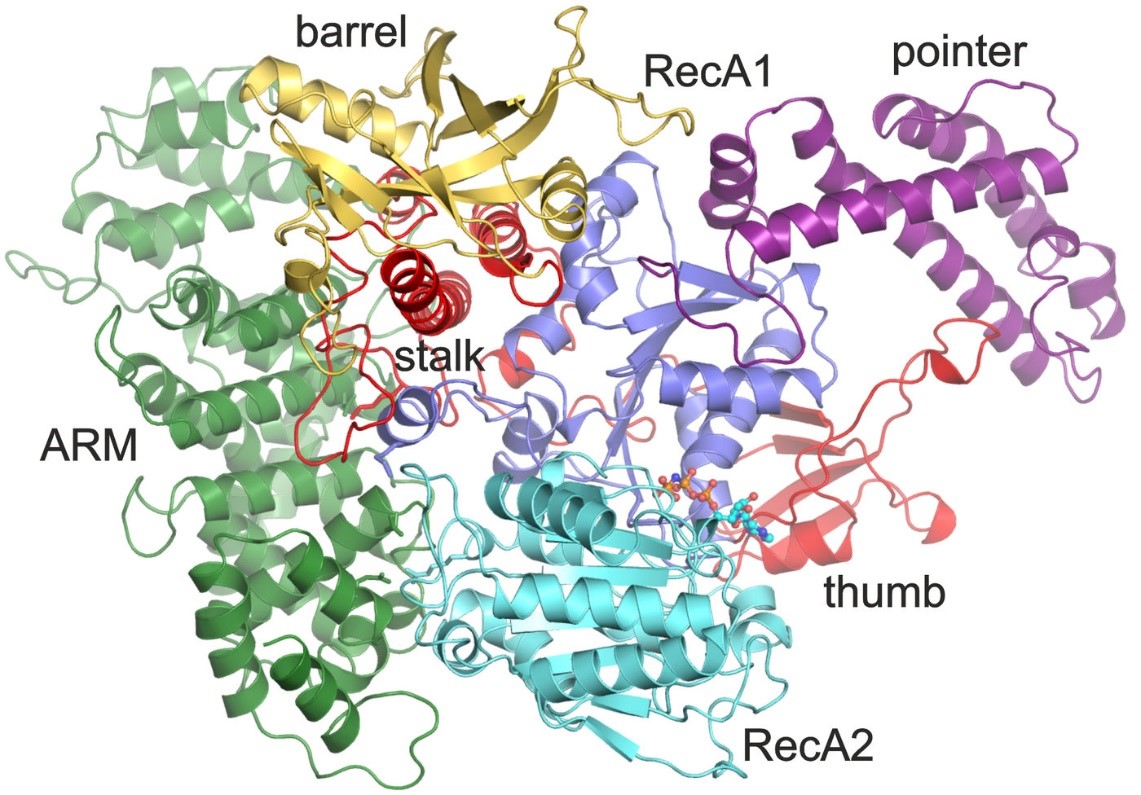
Crystal structure of the RNA helicase Aquarius.
DNA enzymes
Although DNA is known mainly for its capacity to encode information – for instance in the form of genes – DNA molecules that catalyze various chemical reactions have been found in vitro, providing the proof of principle that DNA can act as a catalyst.
Two decades after this discovery, we have determined the first crystal structure of a DNA enzyme. The surprisingly complex fold that these molecules adopt raises questions about the potential structural importance of DNA in the cell, and it enables us to manipulate the molecules' properties for technological applications.
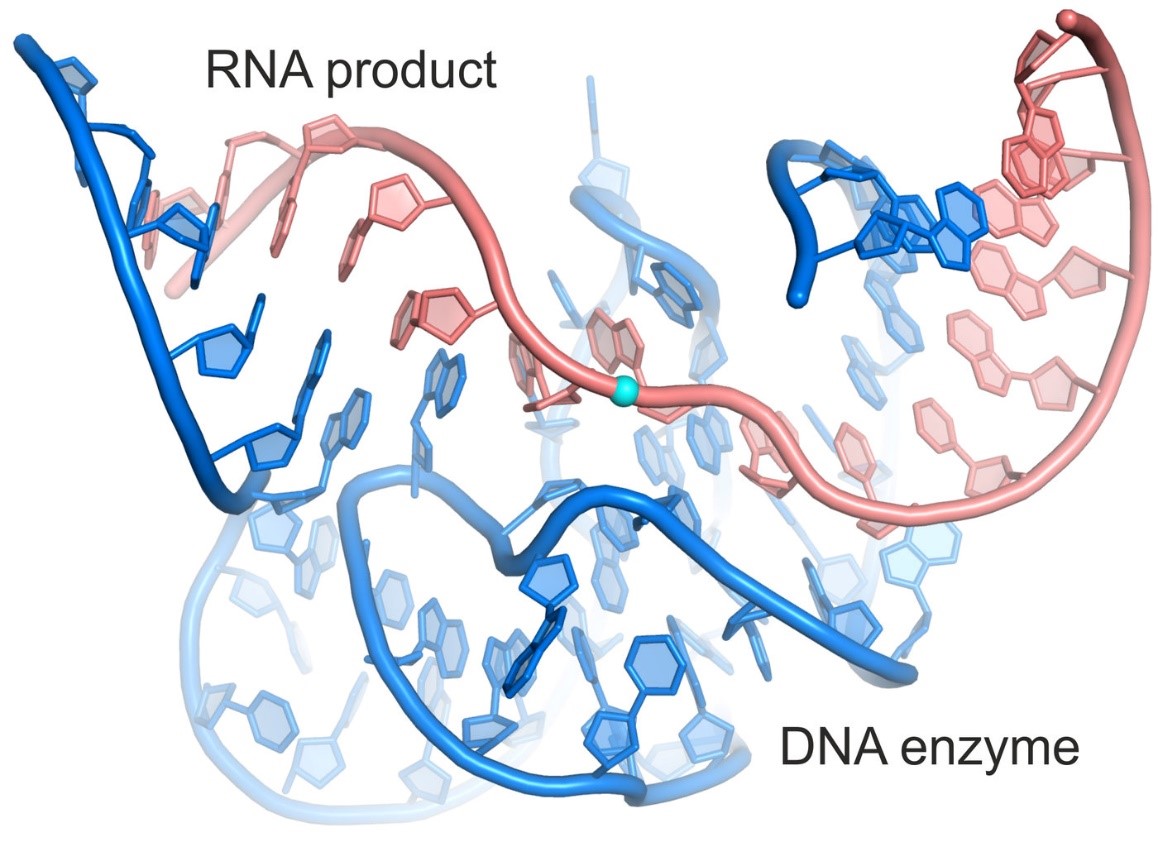
Crystal structure of a DNA enzyme (9DB1) in complex with the ligated RNA product.