CLOSED: The Multiscale Vulnerability Map: Cross-Discipline Multimodal Mining Reveals Colorectal Cancer's Hidden Weak Points
Application closing date: 16/11/25
Project background
Colorectal cancer (CRC) is one of the deadliest cancers worldwide, and patients with a common subtype called MSS tumours face particularly poor treatment options. These tumours are highly adaptable, meaning standard therapies often stop working. To change this, we need to look deeper into the tumour’s biology and uncover its “hidden weak points.”
This PhD project will do exactly that. You will help create the first-ever vulnerability map of colorectal cancer by bringing together huge amounts of data—genomic, proteomic, metabolomic, single-cell, imaging and more—from over 4,300 patients. By analysing this “big picture” with advanced machine learning tools (including our lab’s own iPhenMap platform), you’ll discover therapeutic targets that simply cannot be seen with traditional approaches.
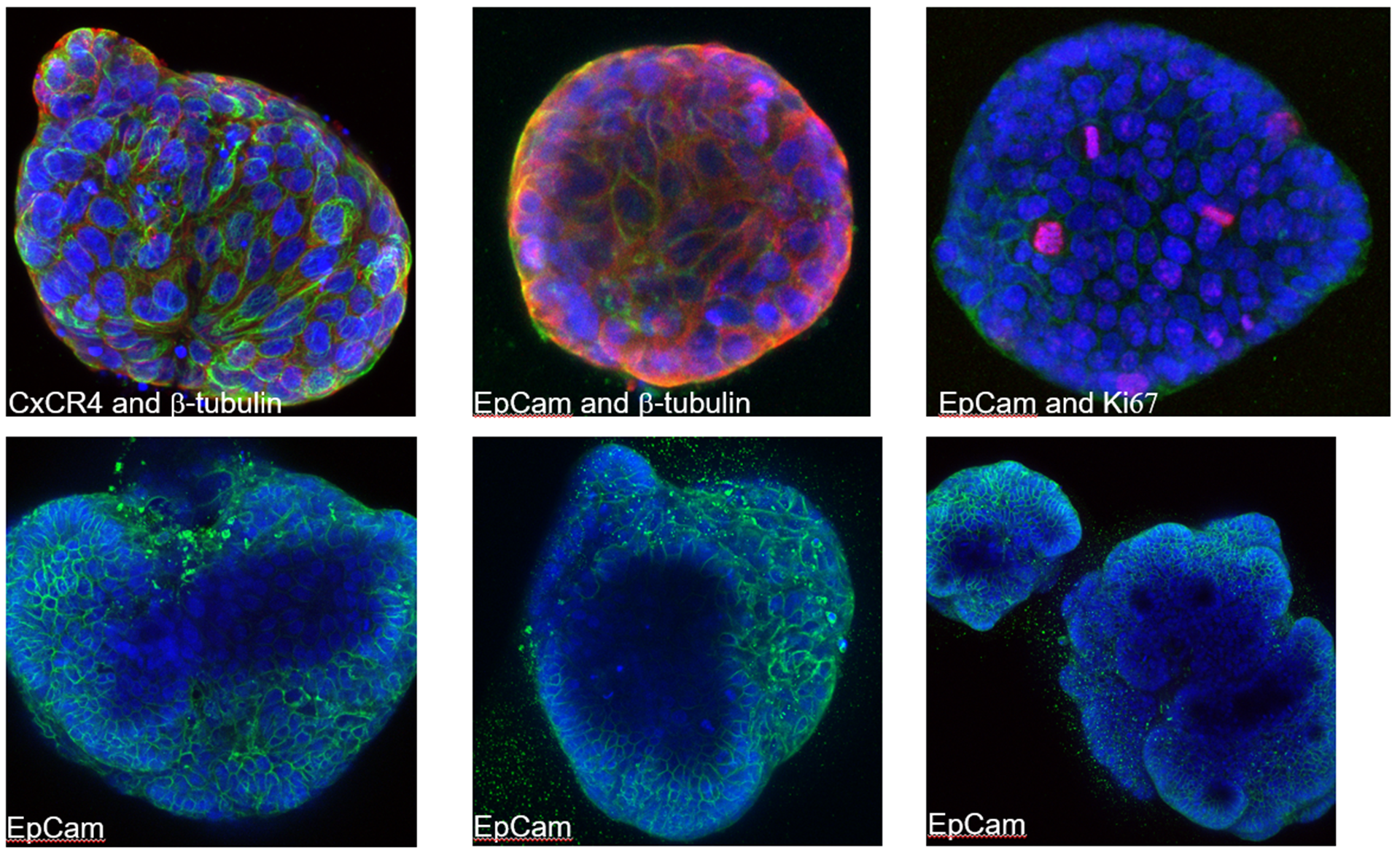
But the project isn’t just about computers and code. You’ll also test these discoveries directly in the lab. Using cutting-edge cancer models—including 3D patient-derived mini-tumours (tumoroids), CRISPR genetic screens, and animal models—you will explore how potential treatments work and identify biomarkers that could predict which patients will benefit most.
A unique aspect of this studentship is the industry collaboration with a Big Pharma, one of the world’s leading pharmaceutical companies. You’ll have access to proprietary drug libraries and clinical expertise and even spend time working in Merck’s research facilities. This will give you a rare chance to experience both academic and industry environments, preparing you for various future career paths.
By the end of this project, you will have built expertise in computational biology, machine learning, experimental cancer research, and precision oncology. Most importantly, you will have contributed to developing the next generation of personalised treatments that could make a real difference for patients with colorectal cancer.
Project aims
- Discover targetable vulnerabilities: Systematically interrogate multiscale, multi-omic datasets using machine learning and iPhenMap integration to identify therapeutic targets invisible to single-platform analyses
- Validate therapeutic potential: Test prioritized vulnerabilities through comprehensive experimental validation using patient-derived 3D tumoroids, xenografts, and syngeneic mouse models
- Develop predictive biomarkers: Create biomarker frameworks from integrated omics-clinical analyses to enable patient stratification and personalized treatment approaches
Further details & requirements
Comprehensive Multiscale Data Integration
This project leverages an extensive collection of multimodal CRC data to build a systematic vulnerability atlas of over 4,000 transcriptomes with established CMS classification.
This represents one of the most comprehensive colorectal cancer datasets ever assembled, enabling analyses at unprecedented scale and depth. The systematic profiling across multiple molecular layers within the same samples allows investigation of cross-modal interactions that single-platform studies cannot detect.
Advanced Computational Framework
We will employ our laboratory-developed iPhenMap platform, which uniquely enables joint integration of multiple omics types with phenotypic covariates in a unified analytical framework. This approach identifies "metavariables" - composite molecular patterns that emerge only when multiple data types are analysed together. Complementary machine learning methods will integrate gene dosage data with functional vulnerability profiles.
Systematic Vulnerability Discovery
Our approach systematically maps therapeutic vulnerabilities through: (1) Re-sampling interaction models combining metabolic and phenotypic features to identify convergent pathway signals across tumor subtypes; (2) Integration with clinical parameters including demographic and prognostic data; (3) Anchoring discoveries within the established consensus molecular subtype taxonomy for clinical relevance; (4) Machine learning-based dependency mapping to identify stress-linked and metabolic vulnerabilities with therapeutic potential.
Comprehensive Experimental Validation
Computational discoveries will undergo rigorous experimental validation using 3D organoids and mouse models. Synergistic drug combinations will be evaluated using dose-response matrices analysed with Bliss and Loewe modelling to identify optimal combination strategies and synthetic lethal interactions.
Biomarker Development Strategy
Predictive biomarkers will be systematically derived from integrated analyses encompassing molecular signatures (replication-stress markers, metabolic profiles, proteostasis modules), phenotypic indicators (anatomical site, perineural invasion, relapse risk, ECOG status), and CMS-anchored classification for immediate clinical translatability.
Expected Outcomes and Clinical Impact
The project will deliver validated therapeutic targets with demonstrated preclinical efficacy, predictive biomarkers enabling patient stratification for clinical trials, and scalable computational frameworks applicable to other cancer types. Multiple discoveries are anticipated to progress toward clinical evaluation, potentially transforming treatment approaches for MSS colorectal cancer.
Scientific outputs will include high-impact publications describing novel vulnerabilities, experimental validation results, and biomarker development, establishing new paradigms for vulnerability-based cancer therapy development.
- Sadanandam, A. et al. (2013) ‘A colorectal cancer classification system that associates cellular phenotype and responses to therapy’, Nature Medicine, 19(5), pp. 619–625.
- Guinney, J. and Sadanandam, A., et al. (2015) ‘The consensus molecular subtypes of colorectal cancer’, Nature Medicine, 21(11), pp. 1350–1356.
- Cancer Genome Atlas Network (2012) ‘Comprehensive molecular characterization of human colon and rectal cancer’, Nature, 487(7407), pp. 330–337.