CLOSED: Elucidation of the molecular basis for the chaperoning of DEAH-box RNA helicases by the oncoprotein and cochaperone NudCD1
Application closing date: 16/11/25
Project background
The NudC domain containing protein 1 (NudCD1, aka CML66 or OVA66) has been identified as a cancer antigen and is an oncoprotein abnormally activated in human cancers [1,2]. It is a member of the NudC protein family, which also contains NudC, NudC-like, and NudCL2. NudCD1 is overexpressed in patients with chronic myelogenous leukemia (CML) and in patient tissues from a variety of cancers including renal, ovarian, lung and colon cancer [2]. Overexpression of NudCD1 in human colorectal cancer (CRC) tissues is associated with advanced disease and metastasis [3]. Knockdown of NudCD1 in CRC lines inhibits cell proliferation, migration and invasion. In addition, silencing of NudCD1 in SKOV3 ovarian cancer cells impaired tumour growth, possibly via attenuation of the phosphorylation of IGF-1R and ERK1/2-HSP27, and inhibited tumour growth in mice bearing SKOV3 tumours [4].
NudCD1 contains a WD40 β-propeller domain and, similar to the other NudC proteins, an HSP20-like p23 domain. The latter suggests a role as a cochaperone of HSP90 [5], but this role and the broader biological function of NudCD1 are poorly understood. However, systematic mapping of the HSP90 cochaperone client network uncovered a strong interaction of NudCD1 with DEAH-box RNA helicases, including the four spliceosomal DEAH-box helicases, DHX16, DHX38, DHX8 and DHX15 [6]. In addition, a characterisation of the three NudCD1 isoforms, identified a near stoichiometric interaction between the nuclear isoform of NudCD1 and DHX15 [7].
The van Montfort group recently co-purified NudCD1 with the helicase domain of DHX38 and confirmed a NuDCD1/DHX38 complex by size exclusion chromatography, dynamic light scattering and mass spectrometry. Moreover, a low-resolution cryoEM map of NudCD1 yielded the first experimental insight into its tertiary structure and a low resolution cryoEM map of the NudCD1/DHX38 complex provided a first glimpse into the interaction of NudCD1 with the DHX38 helicase domain.
Project aims
- Recombinant production of NudCD1 and selected spliceosomal DEAH-box RNA helicases
- Reconstitution, or co expression, of the NudCD1-DEAH-box helicase complexes
- Biophysical, biochemical, and cellular characterisation of the NudCD1-helicase interaction
- Structure determination of NudCD1 and NudCD1/DEAH-box helicase complexes using cryoEM and/or X-ray crystallography
- Uncover the cellular function of NudCD1 in relation to its interactions with DEAH-box RNA helicases and the HSP90 chaperone machinery.
Further details & requirements
Splicing is catalysed by the spliceosome, a large and dynamic protein-RNA complex consisting of five small nuclear ribonucleoproteins (snRNPs) and, in humans, approximately 200 accessory proteins [8]. Major conformational changes are required to enable the two distinct catalytic splicing reactions, and for the subsequent release of the mature mRNA. These extensive conformational changes are mediated by at least eight nucleotide triphosphate (NTP)-dependent RNA helicases belonging to the helicase superfamily 2 (SF2) and include the DEAH-box RNA helicases DHX16, DHX38, DHX8 and DHX15 [9,10].
Structural and mechanistic studies on the different human spliceosomal DEAH-box RNA helicases, and their functional homologs in yeast, have yielded extensive insights into the molecular basis of their RNA-translocations mechanisms [9]. In addition, structural characterisation of landmark spliceosome intermediates during the splicing cycle has vastly increased our understanding of the crucial roles of DEAH-box RNA helicases in remodelling the spliceosome to enable it to deliver correctly spliced mature mRNA [10]. For some DEAH-box helicases, such as DHX15, additional biological roles have been reported and interactions with non-spliceosomal interactions partners have been extensively studied. However, how the DEAH-box helicase are delivered to the spliceosome, and stabilised between disassembly and reassembly of the spliceosome, is poorly understood.
This PhD-project builds on the identification of the HSP90 cochaperone nudCD1 as putative chaperone of the spliceosomal DEAH-box helicases DHX16, DHX38, DHX8 and DHX15, on the separate biophysical and structural studies confirming an interaction between NudCD1 and DHX15, and on the finding of the van Montfort lab that NudCD1 forms a complex with DHX38. The overall aim of the project is to elucidate the molecular basis of the role of NudCD1 in chaperoning the spliceosomal DEAH-box RNA helicases.
NudCD1 has been cloned and successfully expressed and purified in the van Montfort lab and a low-resolution cryoEM map of NudCD1 has been obtained in which the WD40 and p23 domains could be easily distinguished (Figure 1A). As no high-resolution structure of NudCD1 is available a first aim is to determine a high resolution cryoEM and/or X-ray structure of NudCD1, building on the previous work carried out in the group.
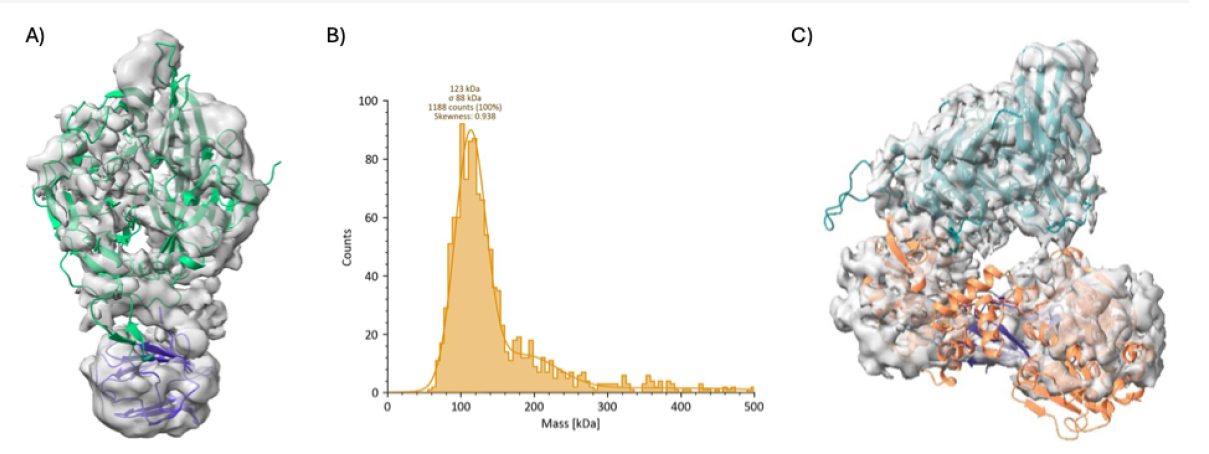
Figure 1. NudCD1 and its interaction with DHX38. A) Low-resolution cryoEM map superimposed with NudCD1 with its WD40 domain in green and the p23 domain in purple. B) Mass photometry plot showing a peak at ~123 kDa corresponding to the DHX38 complex. C) Low resolution cryoEM map showing the binding of NudCD1 to the DHX38 helciase domain (orange) via its WD40 domain (blue).
The second aim builds on preliminary data from the van Montfort lab, which show that co-expressed NudCD1 and DHX38 can be co-purified as a complex confirmed by size exclusion chromatography, dynamic light scattering (DLS), and mass photometry (Figure 1B). Moreover, a low resolution cryoEM map has been obtained which provides a first insight in the binding NudCD1 to the DHX38 helicase domain (Figure 1C). The project also builds on the expertise of the van Montfort lab in generating recombinant DEAH-box helicase proteins [9] and involves recombinant production of the four DEAH-box helicases and subsequent biochemical characterisation, of the different NudCD1/helicase complexes including confirming complex formation using size exclusion chromatography, DLS, and mass photometry. These experiments will be complemented by surface plasmon resonance (SPR) and potentially isothermal titration calorimetry (ITC) experiments to determine the affinity of NudCD1 for the different spliceosomal RNA helicases in order to biophysically characterise the interactions and biochemically evaluate the effects of NudCD1 binding on ATP-hydrolysis and RNA binding of the different helicases.
For the most stable NudCD1 complexes, the next aim is to determine their high-resolution structures using cryoEM (and/or X-ray crystallography) to uncover the molecular details of the NudCD1/helicase interactions. Based on these structures and biochemical/biophysical experiments, mutational studies will be carried out to investigate the impact of disrupting the NudCD1 interactions in vitro or in cancer cells in a biochemical and cellular context measuring helicase ATPase/helicase assays, RNA splicing and viability. The cell-based research will build on the extensive expertise in RNA-helicase biology from the Clarke lab.
The project builds on the extensive in RNA helicase biochemistry and structural biology from Dr Rob van Montfort’s research group and the strong expertise in the RNA biology from the Clarke lab. In addition, the project will benefit from the expertise and ongoing interest of both Drs van Montfort and Clarke in the molecular chaperone field. It is expected that all protein production, biochemical and structural research for this project will be carried out in Dr van Montfort’s group and therefore the student will work at both the Sutton and Chelsea ICR laboratory sites. The cell-based research will be conducted in Dr Clarke group on the Sutton campus.