Image: Graphical abstract from study paper. Credit: Molecular & Cellular Proteomics journal.
Through innovative exploratory work, scientists have identified a new way to study proteins that are involved in the development of the majority of breast cancers.
The research team successfully used a cutting-edge approach to detect most of the proteins known to interact with oestrogen receptor alpha (ERα), which is expressed in about 70 per cent of breast cancers and promotes the development of the disease by increasing cell growth and survival. They also uncovered potential new therapeutic targets in the form of novel proteins that interact with ERα.
Researchers worldwide can now use this approach to help them work towards developing new targeted therapies for ERα-positive breast cancer. They can also use it to study the proteins that interact with key receptors in other types of cancer.
This study was initiated by the Molecular Endocrinology Group at The Institute of Cancer Research, London, in collaboration with the institute’s Functional Proteomics Research Group. It was funded by the Arthur Foundation and Breast Cancer Now and published in the journal Molecular & Cellular Proteomics.
ERα is an essential target
It has long been known that oestrogen has a key role in breast cancer progression. This is largely due to ERα, which is activated by this hormone. Upon activation, ERα crosses into the nucleus of the cancer cell where it interacts with various proteins to increase the number of cancer cells and prolong their survival.
Although scientists have already developed treatments that prevent ERα from binding to oestrogen, many patients find that their cancer becomes resistant to these therapies over time. One of the mechanisms behind this resistance involves abnormal interactions between ERα and the proteins surrounding it.
By understanding the intricacies of these interactions, researchers believe that it will be possible to better understand the mechanisms underlying resistance and to develop more targeted therapies.
Taking a new approach
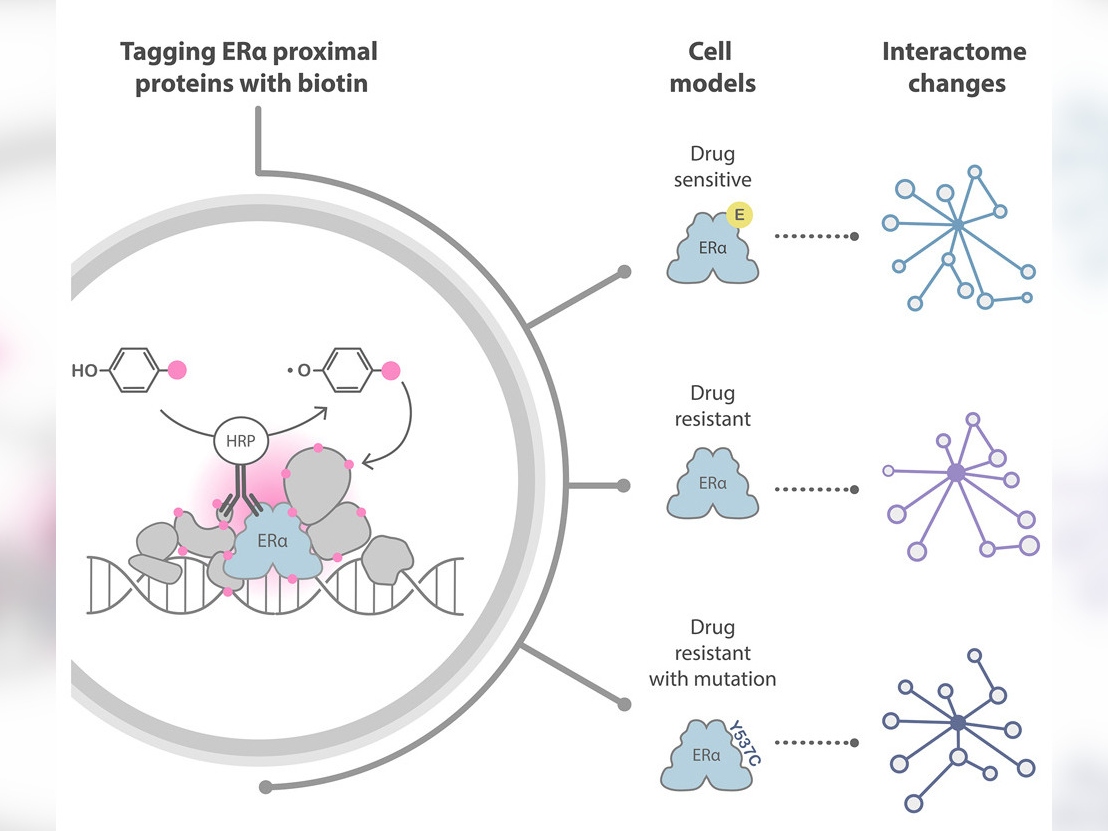
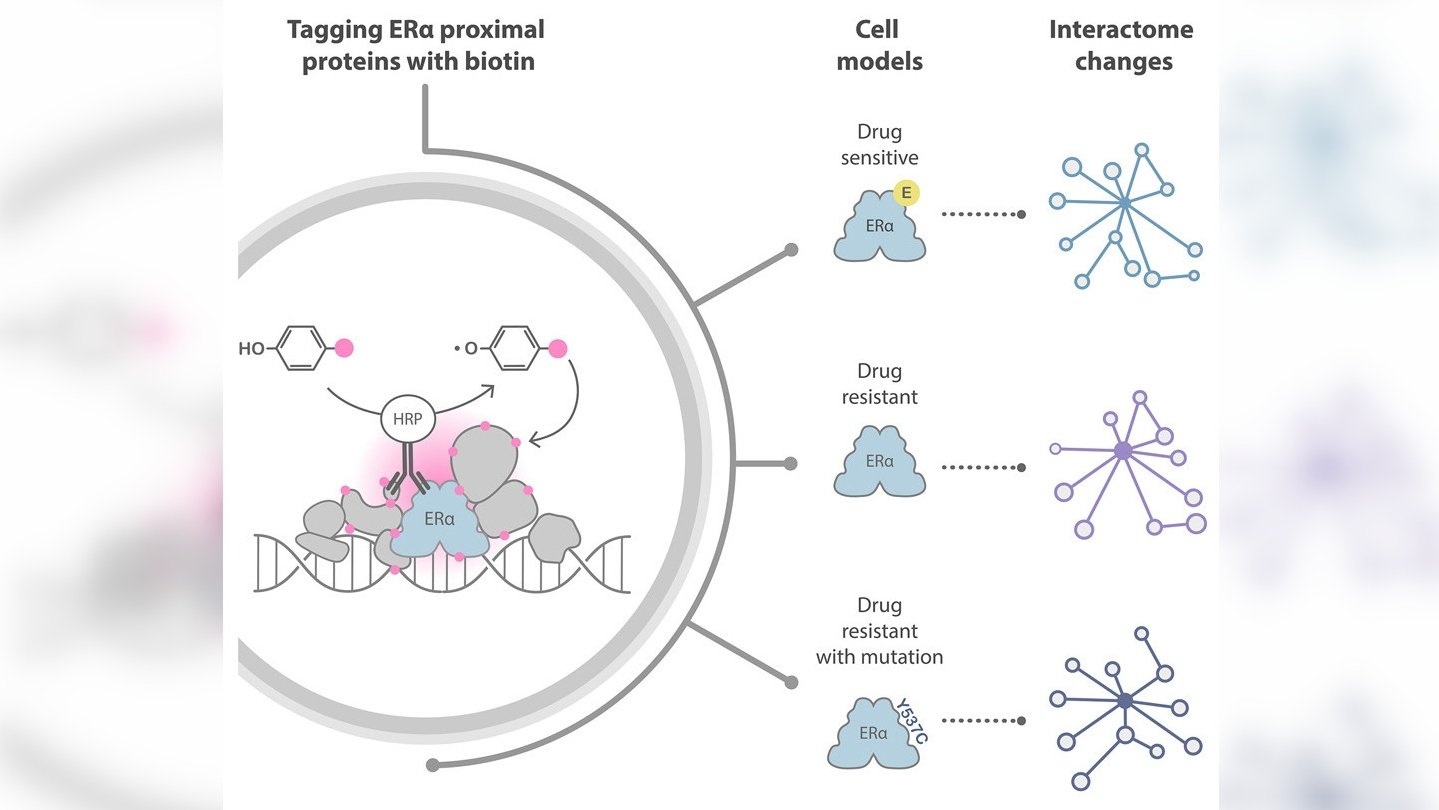
Recently, a novel approach to investigating protein interactions has transformed the protein interaction field. This method is called proximity labelling, and it uses enzymes to label endogenous interaction partners of specific proteins of interest with biotin. An analytical technique called mass spectrometry allows scientists to identify biotin-labelled proteins.
This labelling helps reveal proximal proteins inside cells, but it is not specific enough to reveal the molecular details of how interactions take place. For instance, it cannot distinguish between proteins being next to each other and proteins being directly bound together. However, it is an extremely useful tool for obtaining information about the spatial landscape of proteins.
The team at The Institute of Cancer Research (ICR) decided to try a different technique called biotinylation by antibody recognition (BAR), which uses antibodies joined with enzymes to target the protein of interest, thereby avoiding genetic manipulation of the cells.
For the first experiment, the researchers tested epitope tagging, selecting a short sequence of amino acids (an epitope) that is known to pair with a certain antibody with very high affinity and tagging it onto the proteins of interest so that the antibody could target them. They then used a secondary antibody conjugated with the enzyme to deposit biotin on adjacent proteins in the presence of a biotin substrate and hydrogen peroxidase.
This experiment allowed them to optimise their method for the second part of the study, in which they wanted to target the endogenous protein – protein that has not been genetically modified with the epitope tag. They did this using an antibody able to target the full length of the ERα protein with high affinity.
Confirming BAR’s effectiveness
The mass spectrometry findings revealed that BAR successfully identified 329 proteins known to interact with ERα and revealed 22 new ones.
The researchers were able to use the data from their experiments to identify some of the protein candidates likely to play a part in the development of resistance to treatment. They also refined a list of proteins of interest to 97 that should be prioritised for further scientific investigation.
First author Dr Camilla Rega, a Postdoctoral Training Fellow at the ICR, said:
“This is the first time BAR has been used to investigate ERα’s proximal protein interactions. This technique provides valuable insights into how proteins are spatially organised in the cells and identifies transient interactions that are challenging to detect with traditional approaches.
“Now we have proven that we can use BAR successfully, we plan to apply it to clinical samples from cancer patients so that we can investigate these protein interactions in the context of the disease. A better understanding of these interactions will contribute to the development of targeted therapies for ERα-positive breast cancer and potentially other diseases related to oestrogen signalling.”
Senior author Professor Jyoti Choudhary, Professor of Cancer Proteomics and Head of the Proteomics Core Facility at the ICR, said:
“Our study has provided strong evidence that BAR can be used effectively to further our understanding of ERα’s role in promoting cancer progression and resistance to treatment. It reveals new proteins that are involved in important processes within breast cancer cells.
“This work has opened exciting opportunities in the field to identify new biomarkers and has the potential to gain insights into precision treatment strategies. Our approach can also be used to study the protein interactions that occur with other receptors across cancer types.”