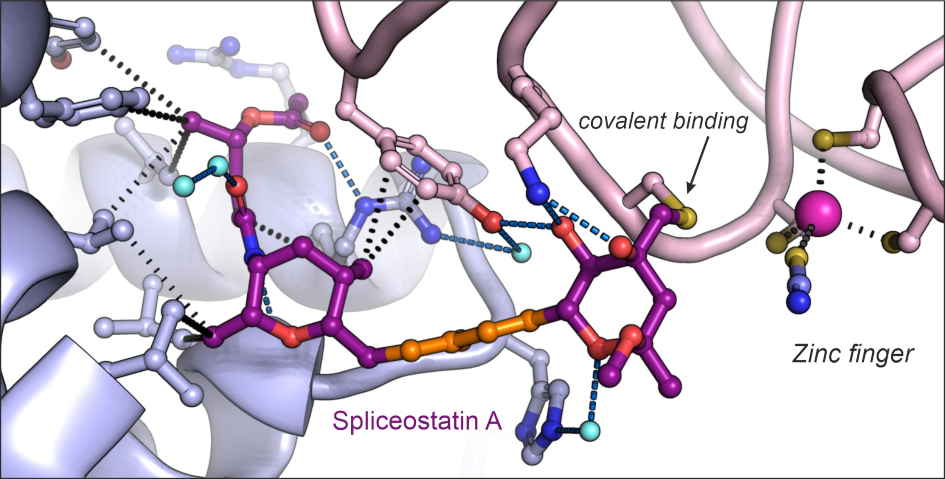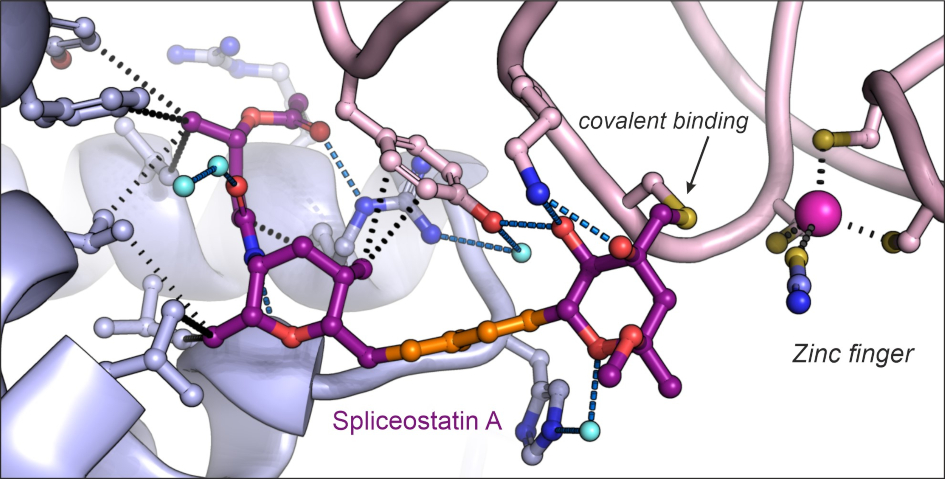
Image: Structure of the antitumor compound spliceostatin A (purple and orange) bound to a spliceosome complex (light blue and pink). Credit: Professor Vlad Pena.
A team of scientists has created a three-dimensional map showing how a small molecule with anticancer properties – called spliceostatin – can promote the killing of cancer cells. This fundamental discovery could ultimately help researchers design future cancer drugs.
The team, from The Institute of Cancer Research, London, revealed how spliceostatin binds the spliceosome – a very large molecular machine that rearranges genetic information within cells through a process known as splicing. While disruption of splicing can lead to cancer, modulation of splicing with spliceostatin can have antitumour effects.
Spliceostatins are representative for a larger class of compounds that have emerged as promising cancer treatment agents due to their ability to target cancer cells. Some of these molecules have even entered clinical trials for various types of cancers.
Mapping out the details
Although spliceostatin was discovered 25 years ago, it has proved challenging to visualise in complex with spliceosomes due to technical limitations.
Recent technological advances in cryo-electron microscopy – a Nobel Prize-winning technique that involves freezing proteins and blasting them with electrons to produce images of individual molecules – have made it possible for researchers to capture the precise molecular details and reconstruct spliceostatin’s 3D shape.
Dr Constantin Cretu, a postdoctoral researcher from the Mechanisms and Regulation of Pre-mRNA Splicing Team led by Professor Vlad Pena, has established the isolation of a human spliceosome complex arrested with spliceostatin.
Using a combination of X-ray crystallography and cryo-electron microscopy, he obtained 3D structures of spliceostatin and sudemycin (another small molecule with anticancer properties) compounds bound to spliceosomal complexes.
Spliceostatin is a powerful tool for dissecting the dynamics of spliceosomes in cells because it can ‘freeze’ the macromolecular machine in its natural state. By capturing the 3D structure of these spliceosomes, the researchers could visualise and understand spliceostatin’s effects on the splicing process in unprecedented detail.
Cancer suppressor
Splicing is important for separating out the information in our DNA that code for proteins from the parts which do not. This process is required to generate messenger RNAs – molecules similar to DNA – that serve as the blueprints for constructing the proteins in our cells.
The relationship between the splicing machinery and cancer has two aspects. On the one hand, genes encoding the splicing machinery are frequently mutated in cancer patients.
The mutations can lead to abnormal splicing patterns in cancer cells, such as the incorrect removal of the protein-coding parts of the RNA and retention of the non-protein-coding parts. Erroneous splicing of the messenger RNA can result in faulty proteins or cause proteins to be wrongly targeted for destruction.
On the other hand, spliceosomes are targets for small molecules with anticancer activities. These compounds, such as spliceostatins, sudemycins or pladienolides can suppress cancer cells by binding and inactivating spliceosomes on certain genes.
This reduces the splicing activity and production of proteins important for cancer cells, ultimately leading to their death.
Remarkably, the ICR team were able to show that spliceostatin and sudemycin can bind to the spliceosome irreversibly, through covalent binding. This is significant because there has been no consensus in the scientific community regarding how these molecules interact.
The prominent study was published in Nature Communications and funded by the German Research Foundation (Deutsche Forschungsgemeinschaft) and the ICR, which is both a research institute and a charity.
Helping to design future anticancer drugs
Study lead Professor Vlad Pena, Team Leader in the Mechanisms and Regulation of Pre-mRNA Splicing Team at the ICR said: “We revealed the first 3D structure of a spliceosome complex bound by spliceostatin, a compound known to supress a wide range of cancer cells.
“This work unveiled in structural terms how this class of compounds modulate or even disrupt splicing in cells. We were surprised to discover that spliceostatin binds covalently to the spliceosome, a fact unknown before despite two decades of research from various groups. Besides the medical implications, we now better understand where and how spliceosomes assemble on the RNA molecules. Our work is consistent with previous observations and unifies a significant amount of scientific research in the splicing field.”
Spliceostatin’s ability to target cancer cells makes it an exciting lead compound for developing anticancer drugs. Ever since its identification as a promising candidate for cancer therapy, various research groups have focused their efforts on producing spliceostatin derivatives or analogues, which are molecularly similar compounds.
These compounds were difficult to synthesise because of the complicated chemistry involved and our limited understanding of their interactions with spliceosomes.
Professor Pena said: “Understanding the molecular and atomic details of how spliceostatin binds to spliceosomes can help inform the design of future synthetic compounds for targeted antitumour treatment. The findings from our study open up new possibilities for identifying and developing more effective anticancer drugs.”