Lego torso by Nick Sawaya from Art of the Brick exhibition. Photo from Pixabay, CC0
Fuelled by both intrigue and narcissism, humans have a long history of trying to understand themselves.
To comprehend something complex, we humans like to take a subject apart and study what we find inside. By reducing an object to its constituent parts, we attempt to understand the whole.
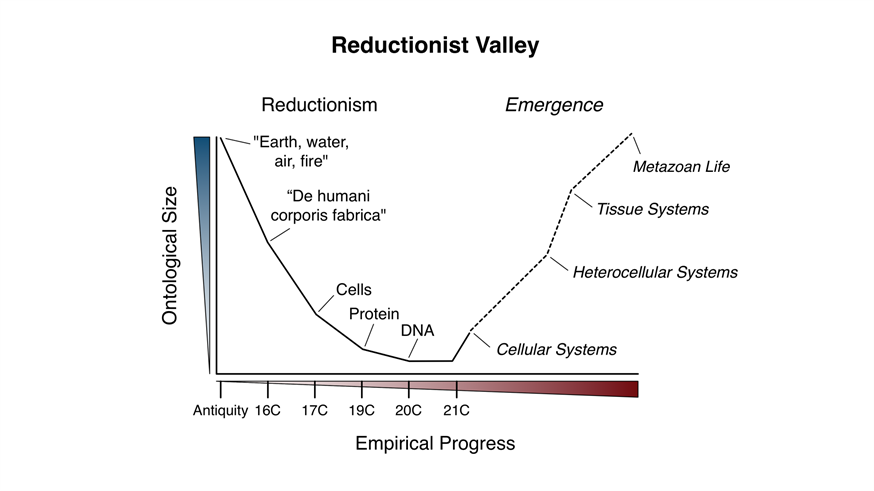
Early endeavours at reducing biology into discrete components were somewhat fanciful. Hippocrates believed humans were formed of ‘earth, water, air and fire’. Aristotle later argued humans were actually composed of ‘hot, cold, wet and dry’.
As empiricism displaced philosophy, we dismantled life with increasing finesse. In the 16th century, the anatomist Andreas Vesalius saw life as a tapestry of bones, arteries, and tissue. He reduced physiology to “De humani corporis fabrica" (The Fabric of the Human Body). In the 17th century, Robert Hooke observed life beyond the naked eye. Peering down his newfangled microscope, Hooke reduced life into even smaller units. He saw little rooms inside our tissue and called them “cells”.
Empowered by technical breakthroughs in physics and chemistry, 19th and 20th century biologists dove deeper and deeper inside these cells. They were found to contain even smaller biochemical constituents such as proteins, lipids, and nucleic acids. Hereditary units — once imagined as nebulous “gemmules” by Darwin — were reduced to a molecule: DNA.
Life had been reduced to the atomic level.
Reductionism
The history of biological science represents a continued narrative of increasing detail towards a molecular depiction of life. All the way from ‘earth, water, air and fire’ antiquity, to our contemporary molecular vista. This process is a classical example of methodological reductionism.
By sequentially teasing life apart, reductionism has provided a detailed, eloquent and beautiful molecular perspective of biology. It’s humbling to consider the thousands of different molecules we confidently describe with atomic precision. The reductionist view of biology is truly spectacular.
The explanatory potential of reductionism is so seductive, modern biological science has become completely addicted to it. It’s where we live. We’ve travelled deep into reductionism and made it our home.
We live in reductionist valley.
Detail ≠ Understanding
Life in reductionist valley is comfortable. It’s a safe place to empirically explore. But like any comfort zone, it's limiting. I believe our residency in reductionist valley is one of the biggest problems facing modern science — and subsequently cancer research.
The core principle of reductionism is that by understanding discrete components of a system, one can extrapolate to understand the whole. Thus, if we keep cataloguing the molecular components of life, we’ll eventually understand how life works.
Unfortunately, this is only true if the discrete components can be re-compiled to form the larger system we are trying to understand. If we can’t assemble the individual components, reductionism becomes a perpetual high-fidelity cataloguing exercise. Just listing the components of a system — not understanding how the system works.
Limited lists
Imagine a pile of Lego bricks.
Some pieces are big, some are small. Some are long, some are wide. Some are traditional bricks, some are specialised. A nearby box displays a completed Lego spaceship — but no instructions on how to build it.
A reductionist can count, measure and document the bricks. They can describe the bricks with absolute accuracy. But ask the reductionist how these bricks combine to build our spaceship, and it becomes less clear. Some bricks look like they might form a wing, maybe others look like the cockpit — but ultimately the reductionist’s list of bricks alone does not contain enough information to build the spaceship. The list is not emergent.
Now imagine our spaceship is a tumour.
A single human cell contains over 20,000 different protein-coding genes, 100,000 protein isoforms, and around 1 x 1010 individual protein molecules. That’s one cell. A small (1 cm3) tumour is composed of approximately 1 x 109 cells. That means a single tumour contains around 1 x 1019 protein molecules.
If a small tumour were built from Lego bricks instead of proteins, it would fill 15 Grand Canyons. Most clinical tumours would be the size of a small country. At this scale, the emergent tumour cannot be captured by a reductionist list – no matter how precise.
The Reductionist Valley — click to enlarge
Escape
Cancer biology has spent a long time making lists in reductionist valley. We’ve made base camp our home and devoted much of our exploring to the documenting of more genes, transcripts and proteins.
The challenge of 21st century biology is not to continue accumulating detail in reductionist valley. Our challenge is to use the reductionist detail we’ve gained to explain higher-order biological systems.
We need to escape from reductionist valley.
But with a lifetime experience in digging down, how can we learn to climb?
Fortunately, wading against the reductionist course is more familiar to cancer biologists than they may realise. For example, each time we ‘functionally validate’ an observation, it often involves traversing up the reductionist slope. A differentially regulated mRNA is ‘validated’ by looking at the protein. We align a ‘lower-level’ reductionist observation with a ‘higher-level’ observation to confirm that the reductionist detail empowers an emergent consequence.
Cellular ‘systems biology’ takes this idea a step further. Just as the 20th century scientists used advances in physics to descend into reductionist valley, a modern systems biologist uses mathematical models of ‘omic-level’ reductionist data to explain emergent cellular phenotypes.
To escape from reductionist valley, we need to continue this ascent.
The next ridge
We’re becoming confident climbers when traversing from genes, to proteins, to cells. The next step is steeper and requires new tools again: we need to understand how reductionist events regulate an emergent population of millions of different cells.
The plural of a cancer cell is not a tumour. Mutated cancer cells exist alongside millions of stromal, endothelial and immune cells — each with their own phenotypic potential. Despite this, we often study tumours as an isolated horde of cancer cells. We sequence thousands and thousands of cancer genomes in the hope that a longer reductionist list will somehow explain the emergence of complex heterocellular pathology.
Yet tumours clearly sit atop reductionist valley. They are emergent heterocellular systems. Should we be surprised we can’t understand them from our view at the bottom?
Explorers
To understand cancer, and ultimately to treat it more effectively, we need to switch our focus from genocentric reductionism to the emergence of heterocellular phenotypes. Fortunately, a new wave of single-cell phenomic technologies is finally providing the tools to achieve this. From single-cell RNA-seq to single-cell signalling mass-cytometry, we are now starting to measure how molecular reductionist events are regulated across huge populations of cells in a tissue. High-dimensional reductionist detail is being used to understand an emergent system.
It’s early days and we have a lifetime of work ahead. The mathematical challenge of integrating such data will be a field in its own right. But finally the tools exist to at least start our ascent.
Scientists have always been explorers — and it's the responsibility of explorers to leave their comfort zone.
The time has come to escape from reductionist valley.