Evolution – the change in a population from generation to generation by mutation, natural selection and genetic drift – conjures up visions of Galapagos beaches and Darwinian finches.
But the concept applies equally well to cancer, as cancer cells undergo mutation, and there is then selection of those cells best able to survive in a tissue ecosystem.
The implications of cancer evolution are devastating – disease progression, spread and eventually death. The process by which evolution occurs is dynamic and highly variable, and its recognition is critical to the successful management of cancer.
Clonal selection of the mutations best able to survive and grow is driven not only by the characteristics of the cells themselves but also by the cells’ immediate microenvironment.
So it is critically important to have methods of recognising characteristic features of aggressive cancer cells or the tissue environments that promote selection of aggressive phenotypes (such as regions of low oxygen saturation).
Modern imaging is providing a means of mapping these kinds of features of tumours with enough accuracy and detail that particular types of image can be used as prognostic markers.
Why imaging?
It is now over a century since Wilhelm Roentgen made the serendipitous discovery of X-rays, allowing us to visualise tissues beneath the surface non-invasively.
Since then clinical imaging systems have themselves evolved at a breathtaking pace, with innovation not just in X-ray based techniques such as computerised tomography (CT) but also in ultrasound, optical imaging, radiofrequency-based techniques within a magnetic field (magnetic resonance imaging, or MRI) and radiolabelled molecular imaging with position emission tomography (PET).
These methods allow imaging down to the molecular scale or up to and including the whole human.
Used in intelligent combination they enable documentation of an array of imaging parameters, so that, for example, a composite map of various features of the tumour ecosystem can be compiled.
When a biopsy is taken, it is restricted to sampling a tiny fraction of the tumour, whereas imaging can non-invasively provide coverage of the whole thing, plus the neighbouring tissues and distant regions to which the cancer may have spread.
This allows interrogation of the behaviour and progression of the tumour within different microenvironments – so, for example, breast cancer cells that have spread to liver may behave and grow differently from those that have spread to the bone.
With imaging, it is also possible to carry out serial examinations at prescribed intervals (although radiation dose and the cost of repeated examinations remain important considerations!)
What can imaging visualise?
An important point to appreciate when using imaging to interrogate a tumour and its microenvironment is the issue of scale.
Current clinical imaging methodologies cannot visualise individual cells or even cell clusters. Instead, they read out in regions of three-dimensional space – known as voxels – of around a cubic millimetre, which within a tumour will contain billions of cells. Often, the voxels displayed on images can be 10 or even 100 times larger.
In comparison, the heterogeneity of the cellular environment of tumours occurs over much smaller distances – just tens of microns. Imaging-derived measurements therefore contribute information over a tumour region that may consist of a variety of microenvironments.
But increasingly imaging techniques are beginning to feed in more detailed information about the development of a microenvironment.
Imaging can, for instance, quantify how many new leaky blood vessels a tumour has sprouted (a process known as neoangiogenesis).
This is done by injecting specific agents into the bloodstream – gas-filled microbubbles for ultrasound imaging, dense iodine-based molecules for CT and weakly paramagnetic agents such as gadolinium for MRI – and mathematically modelling their uptake and washout.
Highly vascular tumours tend to have a poorer prognosis.
Tissue properties such as water diffusion within solid tumours are a surrogate for the density of cell packing and the properties of the extracellular matrix. That means that more restricted diffusion can indicate more aggressive disease. Tumours are also classically hard and tissue stiffness can be quantified using elastography both with ultrasound and MRI.
It is also possible to use imaging to see specific cell populations within individual tumours. We can target receptors expressed on individual cells with fluorescent or radioactive-labelled antibodies in order to view those cell populations using either optical techniques or PET. For example, the androgen receptor status of prostate cancer that has spread to bone can be imaged with radiolabelled dihydrotestosterone, where its overexpression often signals an androgen-insensitive, lethal phase of the disease.
Tumour hypoxia can be interrogated with MRI by capturing the weak effect of deoxyhaemoglobin on the magnetic field in its immediate vicinity.
Alternatively radiolabelled compounds such as imidazoles can be used, which on reduction in hypoxic conditions are trapped within the cell and produce a PET signal.
Finally, metabolic changes in cancer tissue can be probed with both magnetic resonance and PET. The former obtains a metabolite ‘fingerprint’ of atoms visible with magnetic resonance through a method called MR spectroscopy. This can be refined to collect data dynamically – so for example the transformation of one molecule into another, such as pyruvate to lactate, can be monitored to capture the dynamics of metabolism.
We now routinely track tumour activity with PET using radiolabelled glucose as a marker of cellular activity or the uptake of radiolabelled thymidine as a marker of cell proliferation. Increased uptake of these molecules is related to more active and aggressive disease.
An example of multiple MRI parameters available to study prostate cancer is shown in Figure 1 below.
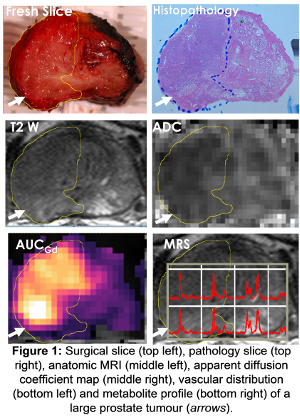
Integrating imaging with measures of evolution
As the imaging signals indicative of poor prognosis tumours are derived from a tumour region of cubic millimetre proportions, they depend on recognising features of the microenvironment.
These are features that either drive the selection of mutant survivor clones (such as hypoxia) or ones that are engendered by them (tumour matrix properties or abnormal vasculature).
Measurement of features such as oxygenation, vascularity, stiffness and water diffusion all help to identify tumours that are likely to grow and spread quickly and have a poor prognosis.
Diffusion-weighted MRI, which characterises the water diffusion properties within tissue and so reflects cell density and matrix properties, is proving to be a particularly valuable prognostic marker in a variety of cancer types.
In cervical cancer, for instance, the quantified apparent diffusion coefficient (ADC) derived from diffusion-weighted MRI is much lower in poorly differentiated tumours than in well-differentiated ones.
A similar trend exists for prostate, breast and bowel cancer. These poorly differentiated tumours will tend to grow faster and have a worse prognosis than better differentiated tumours.
 An excellent human model where features of the tumour microenvironment can be related to growth rate is in low-risk prostate cancer managed by active surveillance.
An excellent human model where features of the tumour microenvironment can be related to growth rate is in low-risk prostate cancer managed by active surveillance.
Patients elect to defer treatment based on a biochemical test called prostate-specific antigen (PSA), imaging and tissue features. They are then monitored with imaging (usually MRI) and with biopsy at intervals if needed.
As the disease often involves more than one tumour in the gland, it is possible to conduct comparative studies between tumours within the same prostate. What has been found is that tumour growth rates are variable with doubling times from 10 months to more than 10 years – and there is now research ongoing to try to relate these differences in growth rates to properties of the different tumours.
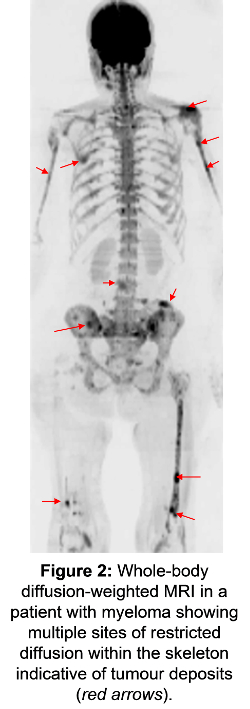
Another good human model system is where multiple sites of secondary tumour exist throughout the body (see Figure 2). Some progress quickly, some less so and some remain stable over long periods.
New whole-body techniques with MRI and PET and the ability to fuse these datasets is allowing us to unravel the metabolic, cellular and vascular features of a tumour and its environment. We can then link these features to variations in disease progression and the development of resistance to treatment.
Looking to the future
Increasingly, imaging is crucial not just in diagnosing a tumour and defining its extent but also in recognising at the outset those tumours that are likely to return or become resistant to treatment.
Mapping a range of imaging parameters (multiparametric imaging) across the tumour provides a composite profile of the tumour microenvironment, currently at a cubic millimetre scale.
The imaging techniques currently of greatest value in providing a range of information that can be fused are MRI and PET. Newer hardware combining these modalities into a single scanner has recently become available and will hugely advance the investigation of tumour biology.
The variation in imaging signals across a tumour (and thus the variability of the tumour microenvironment) is in itself also an important indicator of poor-prognosis disease.
Advances in computational techniques are allowing extraction of "radiomic features" within CT (and ultimately MRI) data – and extraction of 440 such mathematical features in one study was able to decode tumour phenotypes in lung and head and neck cancer and pick out cancers of poor prognosis.
It is possible to use imaging to analyse features indicative of the genetic diversity of a tumour – a critical measure given that it is this diversity which is the substrate of cancer evolution.
Features that indicate genetic diversity, or heterogeneity, across a tumour are strongly correlated with cell cycling pathways, indicating that the most heterogeneous tumours also show increased rates of cell proliferation.
These various techniques could allow early identification of tumours that appear to be rapidly evolving, and are likely to develop resistance to treatment and have a poor prognosis.
It may even be possible to use imaging to pick out treatment-resistant tumour regions that could then receive additional, targeted treatment, preferably under imaging guidance.
The ultimate aim is to use imaging to understand cancer evolution – as a route to ensuring cancer's extinction.