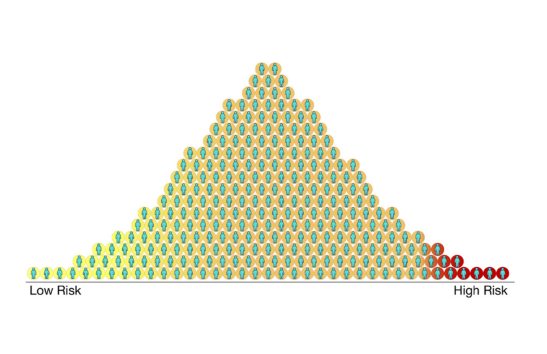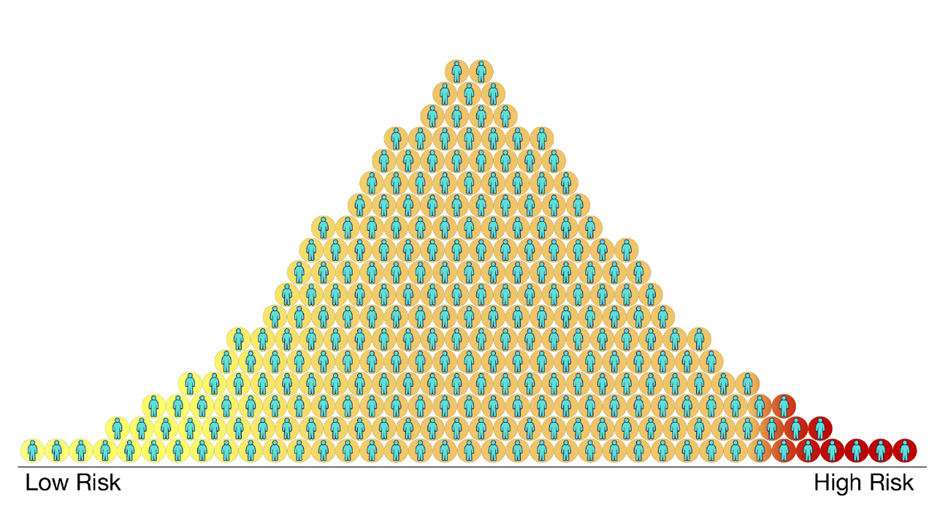
Image: Polygenic risk scores for a disease for different people in a population. Each circle represents a person. Those with higher scores (red) are more likely to get the disease and those with lower scores (yellow) are less likely to get the disease. Most people will fall somewhere in the middle (orange). Credit: Centers for Disease Control and Prevention.
Take a walk through London today and you will spot a new offering shining out from the windows of the city’s swankiest medical clinics: the Polygenic Risk Score. Promising to deliver a ‘personalised estimate’ of your risk of developing cancer, these scores are often described as the key to prevention and early detection – but what are they, and how can they help people to reduce their risk?
So, what’s the score?
We are who we are because of our DNA, a tiny chain of building blocks connected like beads on a string. Each block can be one of four types, represented by the letters A, C, G, and T. When the letters are lined up, they can be read by our body’s machinery as a series of three-letter ‘words’, which together contain all the information we need to grow and survive.
These letters and their corresponding ‘words’ are often grouped into genes, or ‘paragraphs’, which focus on a particular body part or function. When we learn genetics at school, we are taught that there is one gene per characteristic. For example, we learn that there is a gene ‘for’ eye colour, and some of us have the ‘blue’ version, while others have the ‘brown’. However, the reality is much more complex, and there are often several (even hundreds!) of genes that work together to create a certain result.
To understand which parts of our DNA contribute to a particular characteristic, such as eye colour, we find a group of people who do have the characteristic and read letters of their DNA – a process known as genomic sequencing. We then find a group of people who don’t have the characteristic and do the same, and compare the letters across the two groups. We might find, for example, that blue eyed people have an A as the sixth letter of their DNA, while brown eyed people have a T. This variation in the two letters is known as a Single Nucleotide Polymorphism, or SNP, and it tells us both that this particular letter is important for eye colour, and that people with an A are more likely to have blue eyes.
Over time, we can build a library of DNA letters that contribute to a characteristic, which forms the foundation of our polygenic risk score. The more letters a person has that are in that library, the higher their score, and the higher their likelihood of having that characteristic.
Knowledge is power – or is it?
Today, polygenic risk scores have been created for at least 16 types of cancer. Anyone can find out their cancer risk score by visiting a clinic, or ordering a genetic test online. But is it helpful to know?
Searching for a needle in a haystack
It is not always easy to build the library for your polygenic risk score. Groups of ‘letters’ tend to cluster together in chunks across generations, meaning it can be hard to work out exactly which letter is the important one. Once identified, the letter may only have a weak impact on your cancer risk, meaning its influence is hard to discern. And finally, some of the letter changes are very rare, meaning you need to sequence the DNA of lots of people before you spot them.
Geneticists and statisticians have found solutions to many of these problems, and a significant reduction in costs has allowed us to sequence the DNA of more people than ever before. However, most cancers are caused by a combination of genetic and lifestyle factors, meaning that there is a limit to how much cancer risk will ever be explained by DNA.
Lack of representation
The pool of DNA libraries used to build a polygenic risk score is important because it affects which SNPs are included in the score. Unfortunately, most existing DNA libraries are made almost exclusively from the DNA of white Europeans, and don’t reflect the diversity of the global population. This means that many polygenic risk scores don’t perform well for people of non-white ethnicity, limiting their usefulness, and their fairness.
‘High’ and ‘average’ cancer risk may not be that different
For some cancers, the difference in risk between people with ‘high’ and people with ‘average’ polygenic risk score is small. For example, having a high (in the top 5%) polygenic risk score for ovarian cancer translates to an increased lifetime risk, but only from 1.6% to 2.1%1. While this means that a person with a high polygenic risk score is 1.3 times as likely to get ovarian cancer as the average person, their overall risk remains low.
For other cancers, this difference in risk is larger. However, it remains the case that the majority of all cancers will occur in people who are not in the top 1%, 5%, or even 10% of a polygenic risk score. This is simply because there are so few people at high risk, and cancers are so common.
Change isn’t easy
In an ideal world, being told you have a high cancer risk score would come with a series of steps you can take to counteract this genetic predisposition. Perhaps you could exercise more, drink less alcohol, or make sure you attend all your screening appointments. However, big lifestyle changes are difficult to make without support, and for many cancers, accurate screening tests don’t exist. Being informed of a high cancer risk without being able to do anything about it could be a stressful experience, and many may choose to avoid it until better preventative measures area available.
A bright future?
Our DNA is full of information, and while modern technology allows us to read it, we still need to develop our understanding of what it means. Polygenic risk scores sound promising, and may yet turn out to be helpful clinical tools. However, many of these scores are imperfect in their current form, and need thorough testing before entering day-to-day clinical practice. Until then, I will think twice before getting my score calculated!
This piece is shortlisted for the 2022 Mel Greaves Science Writing Prize.
Read more entries from the finalists
_____________________________________________________________________
 Dr Catherine Huntley is a clinical research fellow and public health doctor. She joined the ICR in 2021 as a PhD candidate in Professor Turnbull’s lab, working with NHS Digital to amalgamate and interrogate real-world data to gain insight into prevention and early detection of cancer in people at high genetic risk.
Dr Catherine Huntley is a clinical research fellow and public health doctor. She joined the ICR in 2021 as a PhD candidate in Professor Turnbull’s lab, working with NHS Digital to amalgamate and interrogate real-world data to gain insight into prevention and early detection of cancer in people at high genetic risk.
Prior to joining the ICR, she completed a BA in Human Sciences at the University of Oxford, followed by a degree in medicine at Barts and the London School of Medicine and Dentistry. She then completed her foundation training in London, before commencing specialist training in Public Health Medicine in the same city.
Her research interests include the establishment and curation of national datasets, and the use of real-world data to answer epidemiological questions, with a focus on public health principles such as disease prevention and health equity.