Fragment screening of Checkpoint kinase 2
Checkpoint kinase 2 (CHK2) is a serine/threonine kinase which is crucial in the activation of signal transduction pathways involved in the cellular response to DNA damage caused by external agents. The therapeutic value of CHK2 inhibition is still unclear, but selective CHK2 inhibitors could be potentially beneficial in a variety of contexts.
Professor Michelle Garett (University of Kent, previously at The Institute of Cancer Research) has shown that, while CHK2 inhibition did not potentiate the effect of DNA-damaging chemotherapy, it did sensitize cancer cells to the effects of PARP inhibitors that compromise DNA repair.
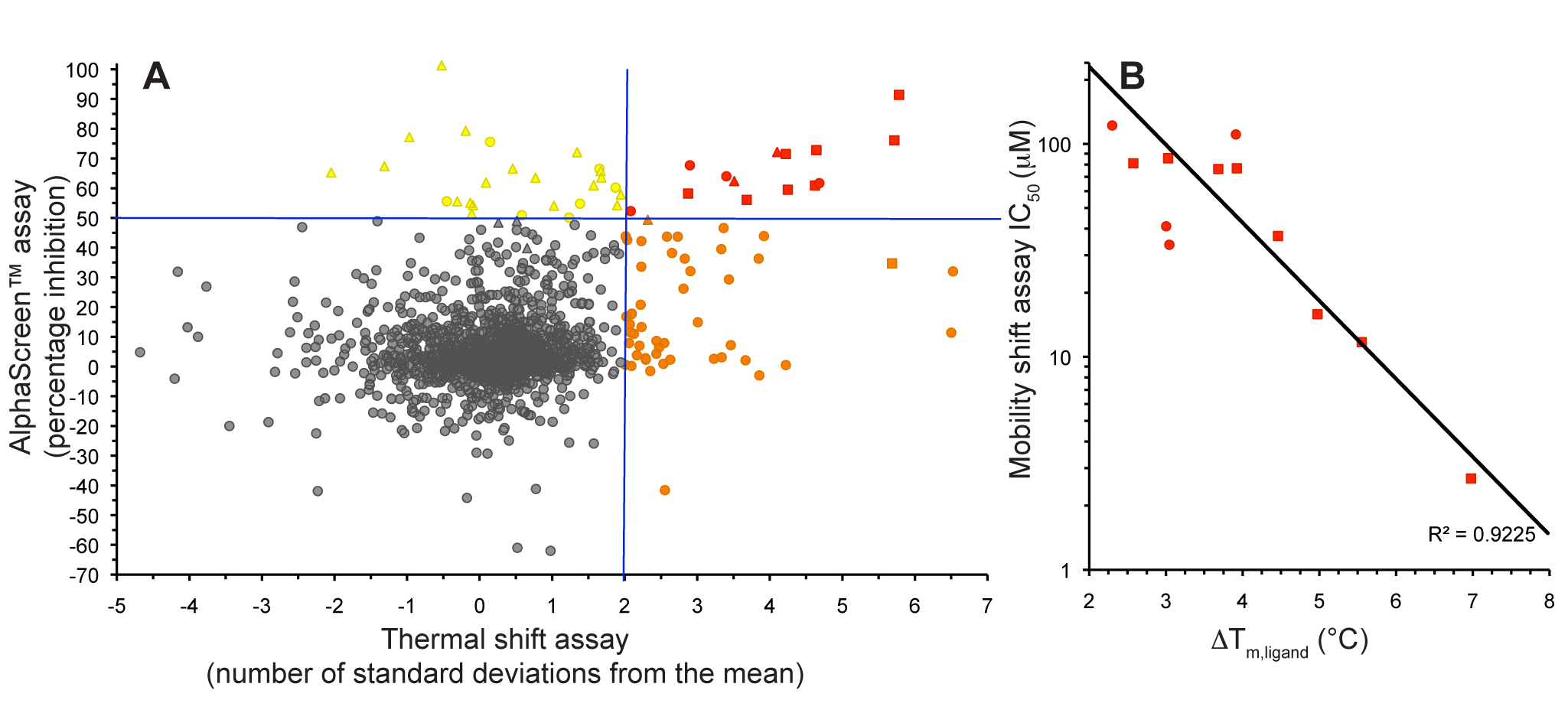
Figure 1. Fragment screening data from biochemical and thermal shift assays. (A) Comparison showing the primary AlphaScreen™ data plotted along the vertical axis as percentage inhibition, and the thermal shift data plotted along the horizontal axis as the number of standard deviations from the mean Tm, ligand for each plate and thresholds displayed as blue lines Hits in AlphaScreen™ and thermal shift are displayed in yellow and orange respectively. Mutual hits are shown in red, inactive fragments are grey. (B) Comparison of the IC50 and DTm, ligand values for the screening hits. Fragments showing interference in the AlphaScreen™ are indicated as triangles. Fragments confirmed in crystallography are shown as squares.
In collaboration with Professors Ian Collins and Michelle Garett we conducted a fragment screen against CHK2 using a combination of a high-concentration Amplified Luminescent Proximity Homogeneous Assay Screen (AlphaScreen™) kinase assay and a thermal shift assay. We showed that the use of these orthogonal techniques allowed efficient discrimination between genuine hit matter and false positives from each individual assay technology (Fig. 1).
We identified a number of chemically different ligand-efficient CHK2 hinge-binding scaffolds that have not been exploited in known CHK2 inhibitors. In addition, the CHK2 crystal structures with a quinoxaline-based fragment and its follow-up compound highlight a hydrophobic area above the hinge region not previously explored in rational CHK2 inhibitor design, but which might be exploited to enhance both potency and selectivity of CHK2 inhibitors.
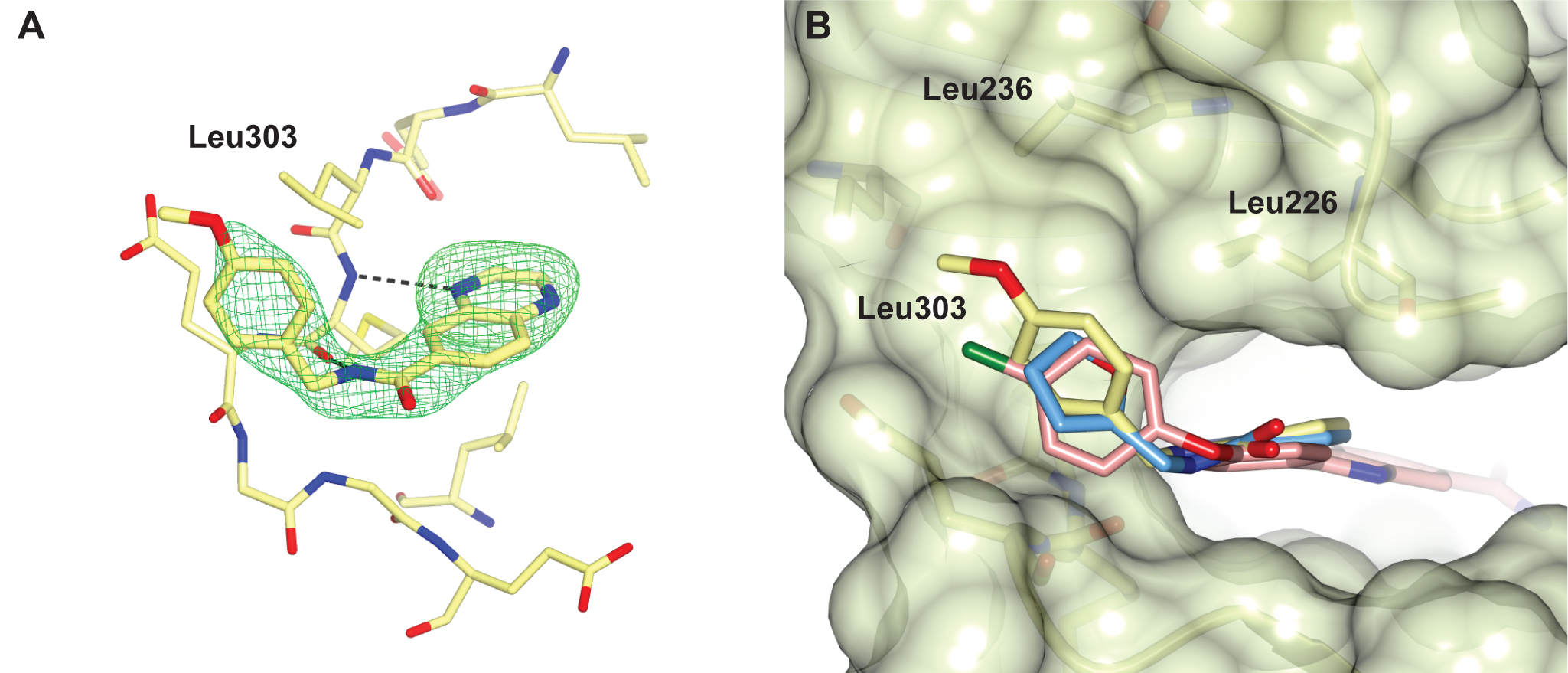
Figure 2 A selectivity pocket above the hinge. (A) Quinoxaline fragment follow-up compound binding to the CHK2 hinge region. (B) Superposition of the quinoxaline fragment (light-blue), its follow-up compound (yellow) and the arylbenzimidazole CHK2 inhibitor (pink, PDB ID code 4A9R), showing the fragments bind in a nearly identical manner with their respective furan and p-methoxyphenyl group binding in a hydrophobic pocket above the hinge, which is also accessed by the chlorophenyl group of the benzimidazole-based CHK2 inhibitor.
A fragment-based approach to target the molecular chaperone HSP70
The 70 kDa heat shock proteins (HSP70s) are an abundant family of ATP-dependent molecular chaperones that are crucial in cellular processes including protein folding, prevention of protein aggregation, modulation of protein complexes, and protein transport between cellular compartments. They have been implicated in several diseases, including cancer and are of significant interest as targets for novel cancer therapies.
In collaboration with Professors Paul Workman and Ian Collins we are applying fragment and structure-based approaches (Fig 3A). HSP70 are extremely flexible proteins (Fig. 3B), therefore a key challenge is to understand how the conformational flexibility of HSP70s affects ligand binding and how it can be exploited in inhibitor design.
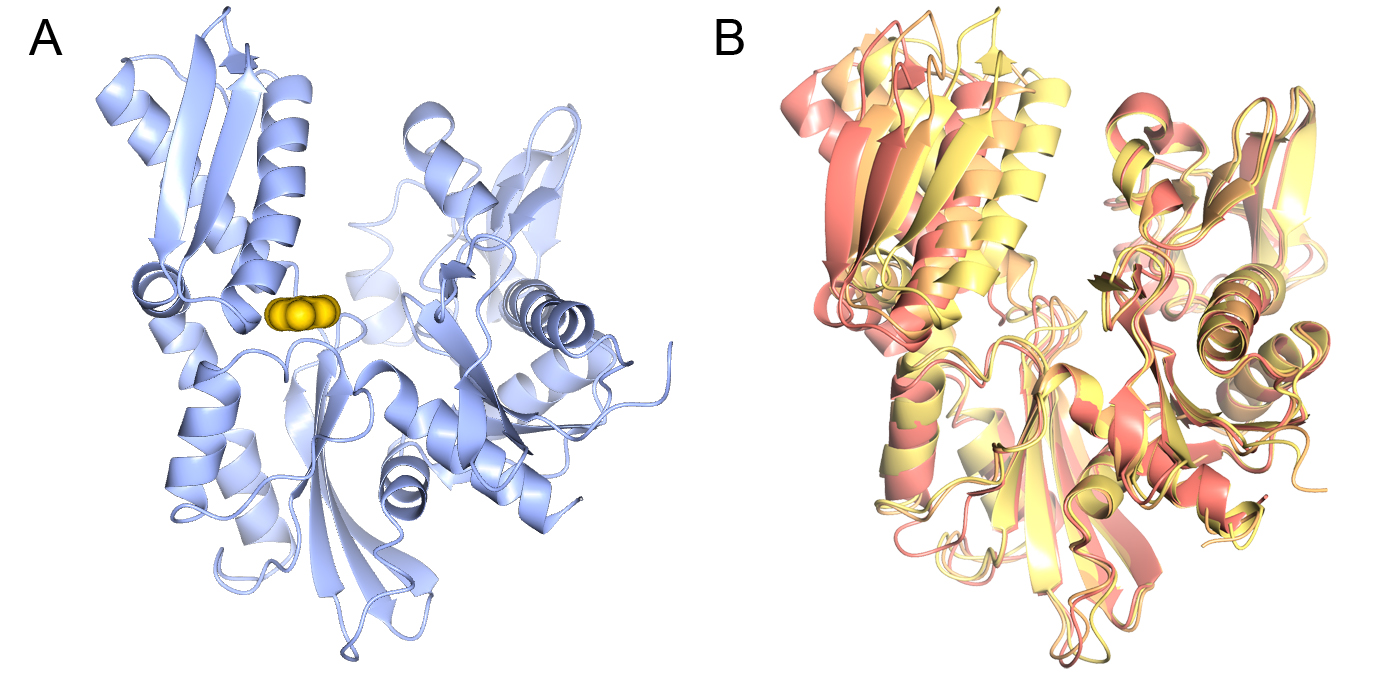
Figure 3 Structure of the HSP70 nucleotide-binding domain. A) A fragment bound in the ATP binding site of the HSP70 nucleotide-binding domain. B) Conformational flexibility on the HSP70 nucleotide-binding domain.