Inhibitors of monopolar spindle 1 (MPS1) kinase
The protein kinase MPS1 is a crucial component of the spindle assembly checkpoint signal and is aberrantly overexpressed in many human cancers. MPS1 is one of the top 25 genes overexpressed in tumors with chromosomal instability and aneuploidy; PTEN-deficient breast tumor cells are particularly dependent upon MPS1 for their survival making it a target of significant interest in oncology.
We are collaborating with Professors Spiros Linardopoulos, Julian Blagg and Dr Swen Hoelder and the CRT Pioneer fund LP to discover small molecule inhibitors of MPS1. Using a high throughput screen of an in-house kinase-focused library combined with a structure-based drug discovery approach, we have discovered CCT251455, a potent, selective and orally bioavailable chemical tool for MPS1. Interestingly, CCT251455 stabilizes an inactive conformation of MPS1 with the activation loop ordered in a manner incompatible with ATP- and substrate peptide–binding (Fig 1).
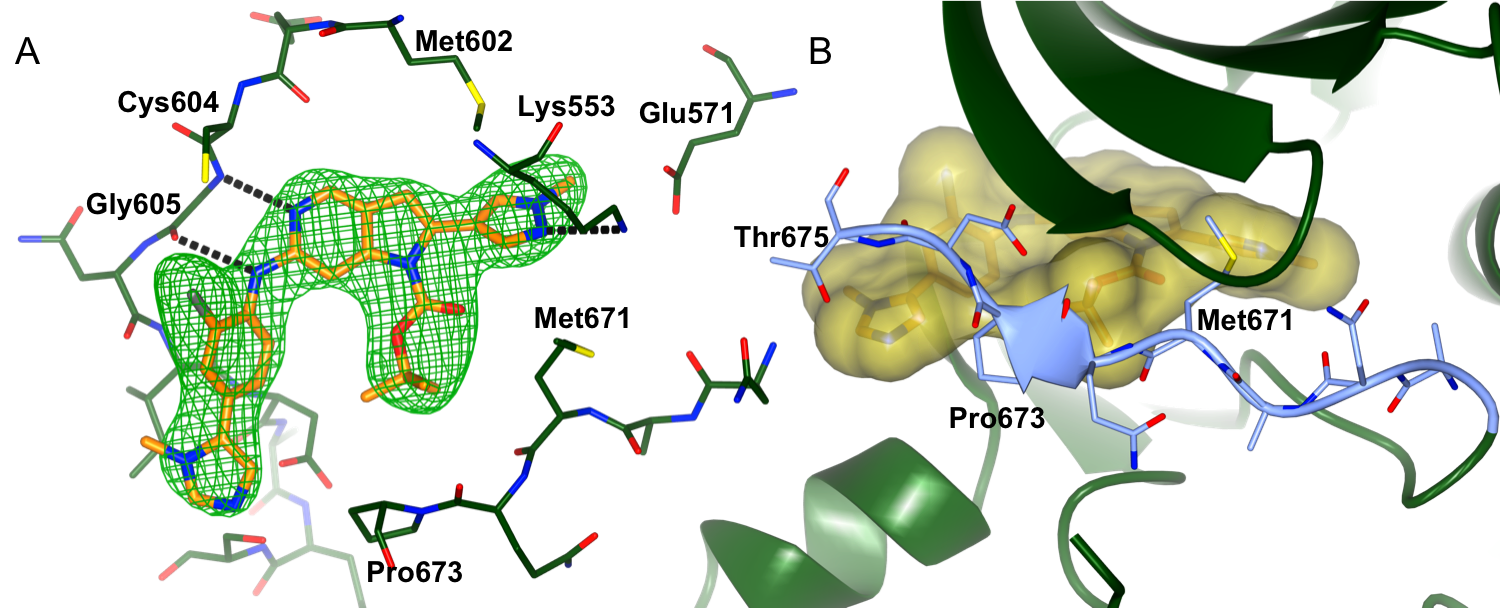
Figure 1: Binding of CCT251455 to MPS1. (A) Binding of CCT251455 to the hinge region of MPS1. (B) CCT251455 stabilises and unusual conformation of the activation loop (light blue).
Inhibitors of the KDM4/5 JmJC histone demethylases
Histone demethylases are involved in the regulation of the methylation status of lysine residues on the N-termini of DNA-bound histones, a process increasingly implicated in cancer. The JmjC histone demethylase proteins are a family of Fe(II) and 2-oxoglutarate (2-OG)-dependent enzymes which convert trimethylated lysine substrates to their dimethylated forms. The KDM4 JmJC demethylase subfamily consists of six members.
In collaboration with Professor Julian Blagg and the Structural Genomics Consortium (SGC, Oxford) we aim to discover novel and inhibitors selective for the KDM4 subfamily. From a high-throughput screen followed by a structure-based drug discovery approach we have discovered potent inhibitors selective for the KDM4 and KDM5 histone lysine demthylases (Fig. 2).
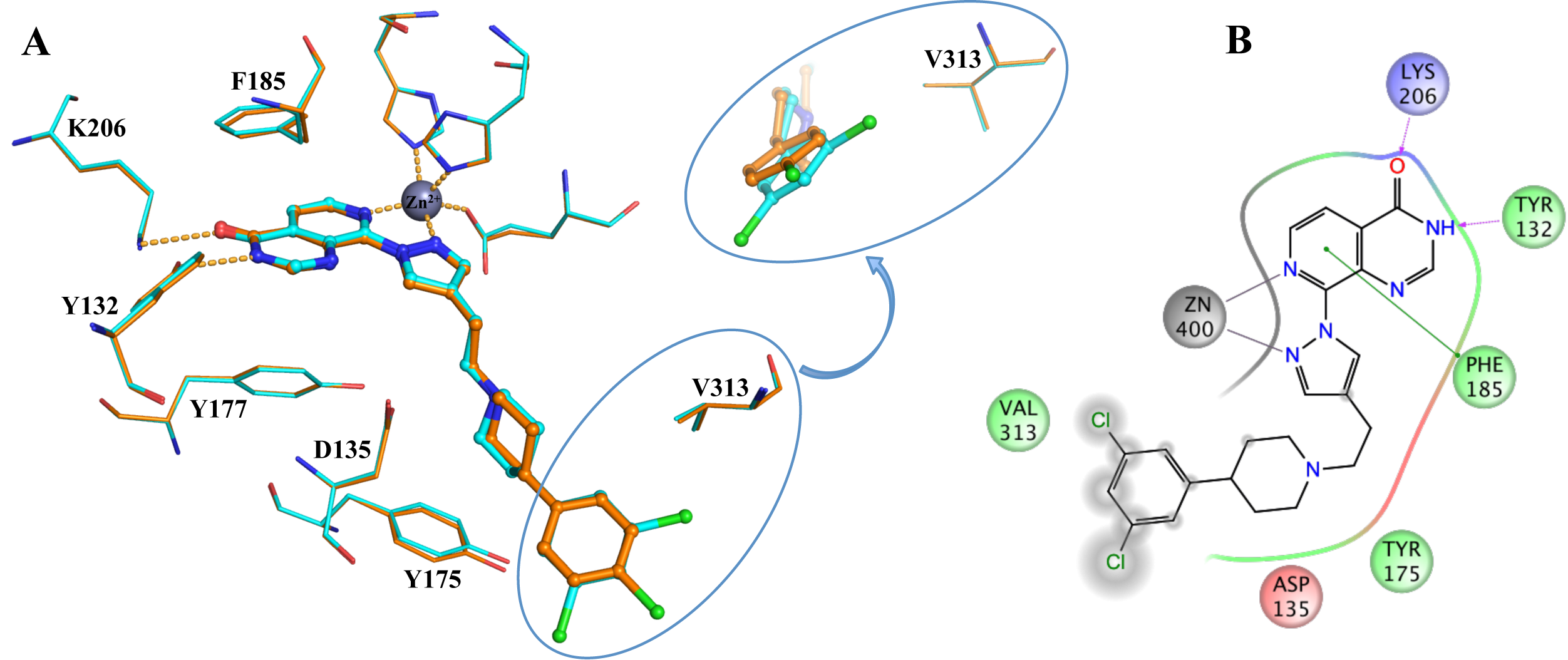
Figure 2: Binding of KDM4/5 inhibitors to KDM4A. A) Superposition of two KDM4/5 inhibitors (blue/organge) bound to KDM4A highlighting key interactions with the protein. B) 2D interaction diagram showing the interactions of the most potent inhibitor (orange) with KDM4A.
AGC kinase inhibitors
Diazaspirocyle-based PKA inhibitors
One of the best protein kinase in the AGC kinase family is the cAMP-dependent protein kinase A (PKA). As part of a collaboration with Professor Ian Collins to evaluate substituted diazaspirocycles as scaffolds to probe the ATP-binding site of protein kinases, we determined high-resolution protein-ligand structures of several diazacpirocycle-based inhibitors (Fig 1).
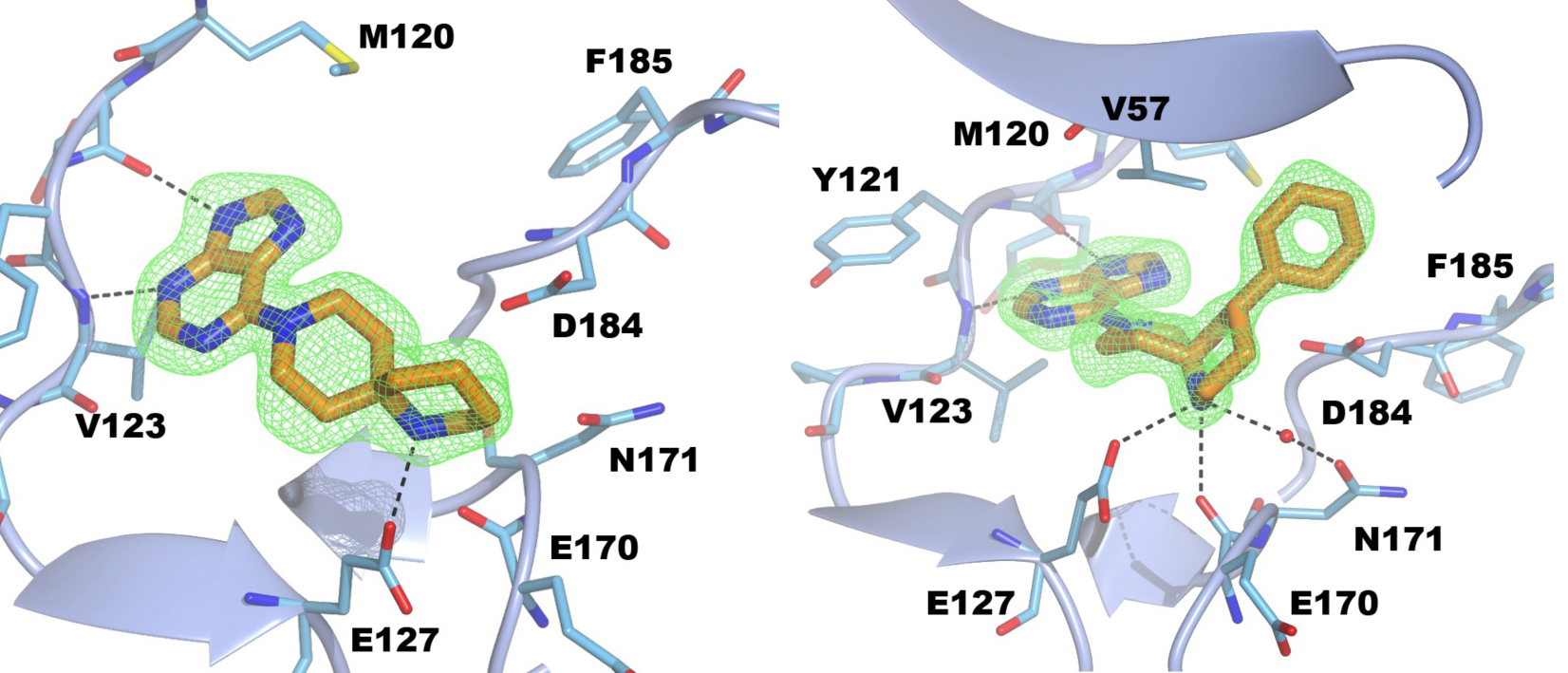
Figure 3: Crystal structures of PKA in complex with two different spirocycle-based inhibitors.
S6K inhibitors
The 70 KDa ribosomal S6 kinases (S6K) RPS6KB1 (S6K1) and RPS6KB2 (S6K2) are key effectors of PI3K/mTOR-regulated signalling, and have been implicated in a variety of human diseases including diabetes and cancer. They belong to the family of AGC kinases and are highly homologous with a sequence identity of 83% in their catalytic domains. We are not pursuing the discovery of S6K inhibitors for the treatment of cancer, but our crystallography efforts provide an example of the use of chimeric proteins as structural surrogate systems in structure-based drug discovery. Currently, no apo-structures of S6 kinases are avaliable due to a susceptibility of the apo-enzyme to aggregation. Reported S6K1 structures are protein-ligand complexes with the pan-kinase inhibitor staurosporine, making the system unsuitable for iterative protein-ligand crystallography to study ATP binding site inhibitors.
To generate a robust crystal system suitable for the generation of high-resolution protein-ligand structures to facilitate S6K inhibitor design, we developed a S6K1 chimeric protein based on the catalytic subunit of the closely related AGC kinase PKAα (PKA, Fig. 4).
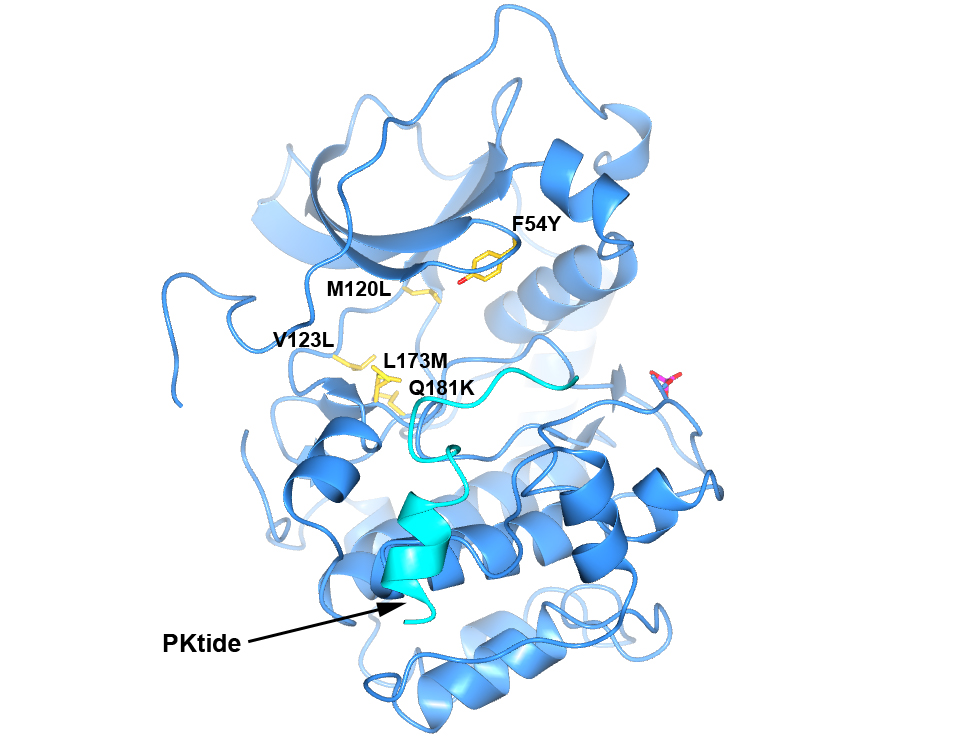
Figure 4. Structure of PKA with the five mutations in and near the ATP-binding site highlighted in yellow. The PKtide substrate inhibitor is shown in light blue
S6K1 and PKA share a sequence identity of approximately 33% in their kinase domains and their ATP-binding sites are nearly identical. Our PKA-S6K1 chimera was created by mutation of five residues in or near the ATP-binding site (F54Y, M120L, V123L, L173M and Q181K) that differ between the two kinases. Using this chimera we could obtain high-resolution structures of two inhibitor series identified in our high-throughput screen carried out in collaboration with Professor Keith Jones and scientists from Newcastle University (Fig. 5).
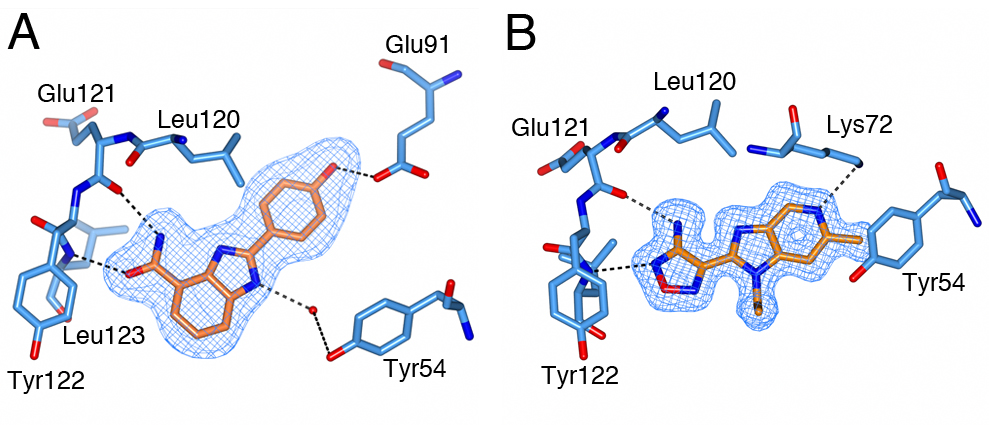
Figure 5. S6K inhibitors bound in the PKA-S6K1 chimeara. A) High resolution structure (2.2Å) of a carboxamidobenzimidazole HTS hit bound in the PKA-S6K1 ATP binding site. B) High resolution structure (1.6Å) of a benzimidazole oxadiazole S6K inhibitor bound in the PKA-S6K1 ATP binding site.