Synthetic lethal approaches to cancer treatment
Our main research programmes, funded by Breast Cancer Now and Cancer Research UK, focus on understanding how to exploit the genetic changes that occur in tumours to develop new treatments. The genetic changes that drive cancer growth can paradoxically result in weaknesses not found in normal cells, which can represent targets for new cancer therapies.
For example, we found that cells lacking the common breast cancer tumour suppressors BRCA1 or BRCA2, which repair DNA through homologous recombination (HR), were remarkably sensitive to inhibition of PARP1, a protein involved in a separate form of DNA repair. This discovery established a paradigm for targeting DNA repair proteins in HR-deficient cancers. The effectiveness of this targeted therapy in patients has resulted in FDA approval of PARP inhibitors for the treatment of BRCA-mutant breast, ovarian and pancreatic cancers.
To identify further synthetic lethal interactions, our lab employs the latest genome engineering technology to manipulate cells at a genome-wide level. From transposon mutagenesis and siRNA/shRNA transfection to modern CRISPR-based mutagenesis, activation, and inhibition screens, we regularly incorporate new technology into our lab. We use these to knockdown, knockout, overexpress or repress every possible gene to search for unique dependencies of some of the most common cancer mutant backgrounds.
Through this work, we have identified synthetic lethal candidates for a series of cancer-associated genes affecting diverse cellular processes such as cell cycle regulation, transcription, RNA splicing, cell adhesion, and chromatin structure which has led to additional animal studies and clinical trials.
We have continued investigating the vulnerabilities of HR-defective cancers and have also begun studying how the unique genetic profile of a tumour may contribute to drug resistance in breast cancer patients.
We are always interested to hear from potential postdoctoral researchers or graduate students with an interest in these subjects – please use the email links on our Team page to get in touch.
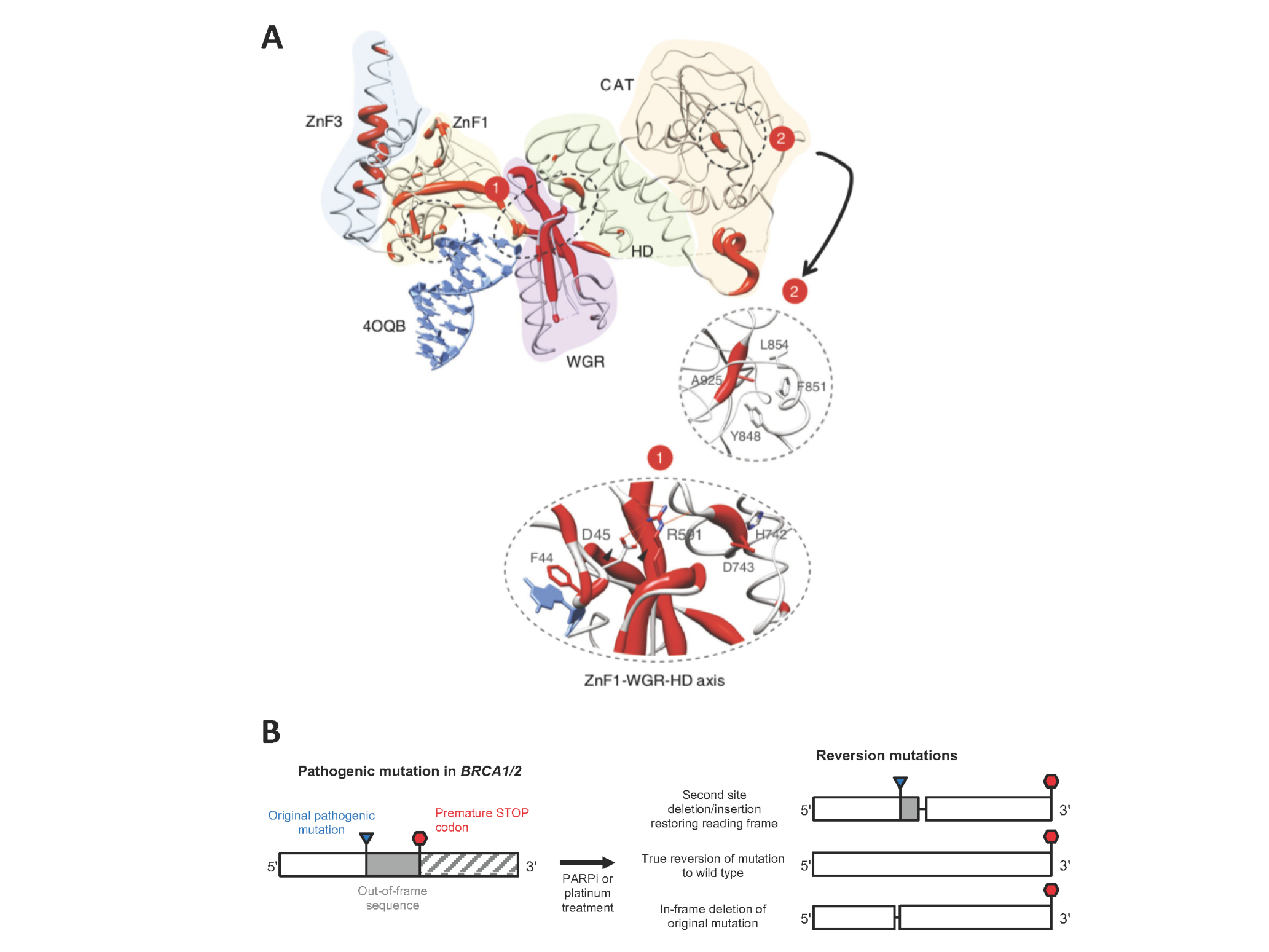
PARP inhibitor resistance. A) Map of resistance-causing mutations identified in a focused PARP inhibitor resistance screen on the crystal structure of PARP1 bound to DNA (Figure from Pettitt et al. Nature Communications 9:1849 (2018)). Regions with high frequency of mutations causing resistance are shown in red. B) Common mechanisms of restoration of function of mutant BRCA genes via reversion mutations.
Mechanisms of PARP inhibitor resistance
PARP inhibitors are now in routine clinical use for a number of indications, including BRCA1/2 mutant breast, ovarian and pancreatic cancer, as well as in clinical trials in further settings. Although PARP inhibitors have demonstrated efficacy in these tumour types, development of resistance has been observed.
Understanding the basis of this resistance can teach us more about the mechanisms of action and determinants of sensitivity to PARP inhibitors, and modelling resistance allows us to propose further therapeutic strategies for trials in patients that develop resistance. We approach this both through lab-based approaches such as genetic screens and by analysis of biopsy material from patients who develop resistance in the clinic.
Our previous genetic screens have enhanced our understanding of how PARP inhibitors kill cells. We found that most PARP inhibitor cytotoxicity depends on PARP1 itself, and that cells with loss of function mutations in PARP1 – including in the DNA binding domain – become extremely resistant to PARP inhibitors (Pettitt, Krastev et al. Nature Communications 2018, Figure 1A).
This supports the “trapping” hypothesis of PARP inhibitor action, where lack of PARP1 activity is not the main cause of cell death when cells are exposed to PARP inhibitors. Rather, inactive PARP1 becomes stuck at sites of damage in the presence of inhibitor, unable to dissociate from the DNA and causing a more toxic form of DNA damage. We are continuing to study how cells deal with “trapped” inhibited PARP1 as a route to further understand and improve PARP inhibitor treatments for cancer.
We also carry out genome-wide screens in a variety of cancer cells to identify new determinants of PARP inhibitor sensitivity and resistance. Such screens can identify new components of the DNA damage response that are involved in the processing of PARP-inhibitor induced DNA damage, a recent example being the Sheildin complex (Noordermeer et al. Nature 2018).
As well as being guided by hypotheses arising from our laboratory work, we also analyse biopsies from patients that become resistant to PARP inhibitors in the clinic. One commonly observed mechanism of resistance in the clinic is reversion mutations, where the mutant BRCA1/2 allele acquires a second mutation that compensates for the pathogenic mutation by restoring the open reading frame (Figure 1B).
We maintain a database of these mutations (reversions.icr.ac.uk) and are characterising how they arise, how they could be prevented and how cells with reversion mutations could be targeted for treatment.
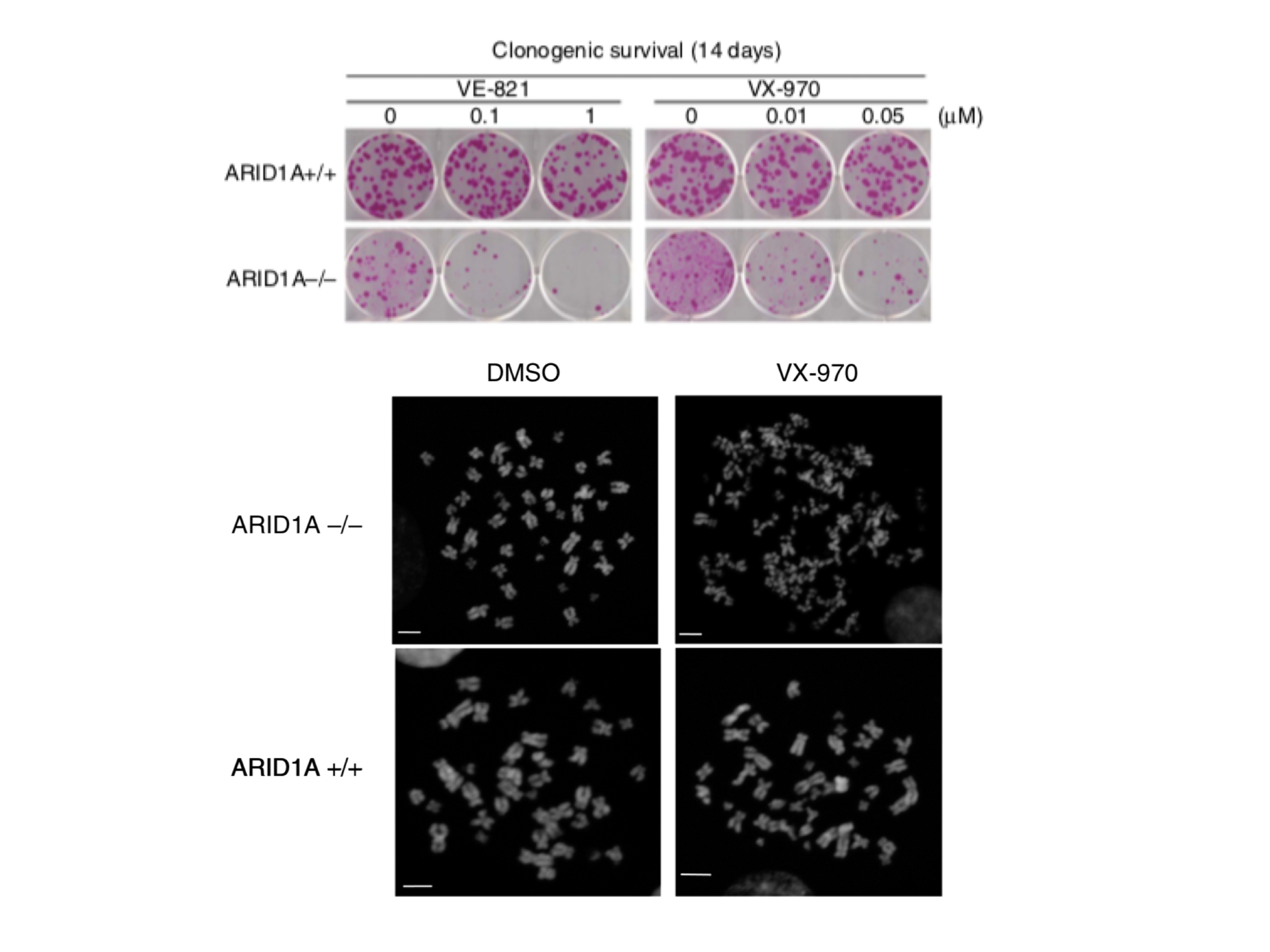
Upper panel: Sensitivity to ATR inhibitors (VE-821 and VX-970) in cells with mutations in ARID1A.
Lower panel: Chromosome “shattering” in cells with ARID1A mutations upon treatment with the ATR inhibitor VX-970 compared to solvent control (DMSO). Figure from Williamson et al. Nature Communications 7:13837 (2016)
Targeting SWI/SNF deficiencies in cancer
Building on our identification of synthetic lethality between BRCA1/BRCA2 mutations and PARP inhibitors, we have focussed on identifying synthetic lethal interactions in other common cancer contexts.
One of the most common recurrent events observed in cancer is the loss of ARID1A function. ARID1A is a critical component of the SWI/SNF complex which is instrumental in regulating chromatin structure. Considered as a complex, mutations in genes encoding SWI/SNF components are found in around 20% of overall cancer cases.
We have shown that ARID1A mutations lead to defects in TOP2A and cell cycle control, sensitising to a class of drugs known as ATR inhibitors (Williamson et al., Nature Communications 2016, Figure 2). Furthermore, we have been able to demonstrate that synovial sarcomas expressing SS18-SSX fusion genes are also profoundly sensitive to ATR inhibition (Jones et al., Cancer Research 2017). Of particular interest, these oncogenic fusion genes disrupt composition of the SWI/SNF complex, reinforcing the idea that SWI/SNF mutant cancers may be targetable by exposure to ATR inhibitors.
These studies and others in the field have encouraged establishment of clinical trials of ATR inhibitors in SWI/SNF-defective cancers, such as the ATARI trial being conducted at the Royal Marsden.
Work is ongoing in our group to inform further applications of ATR inhibitors in two complementary ways. We aim to discover additional synthetic lethal contexts and potential sensitisers to ATR inhibitors that may increase the proportion of cancers that can be effectively treated by these drugs. Conversely, we are also aiming to identify and pre-empt potential resistance mechanisms to these drugs by identifying likely secondary mutations and testing effective therapeutic drug combinations pre-clinically. We are approaching these questions both through CRISPR screens and hypothesis driven experiments.
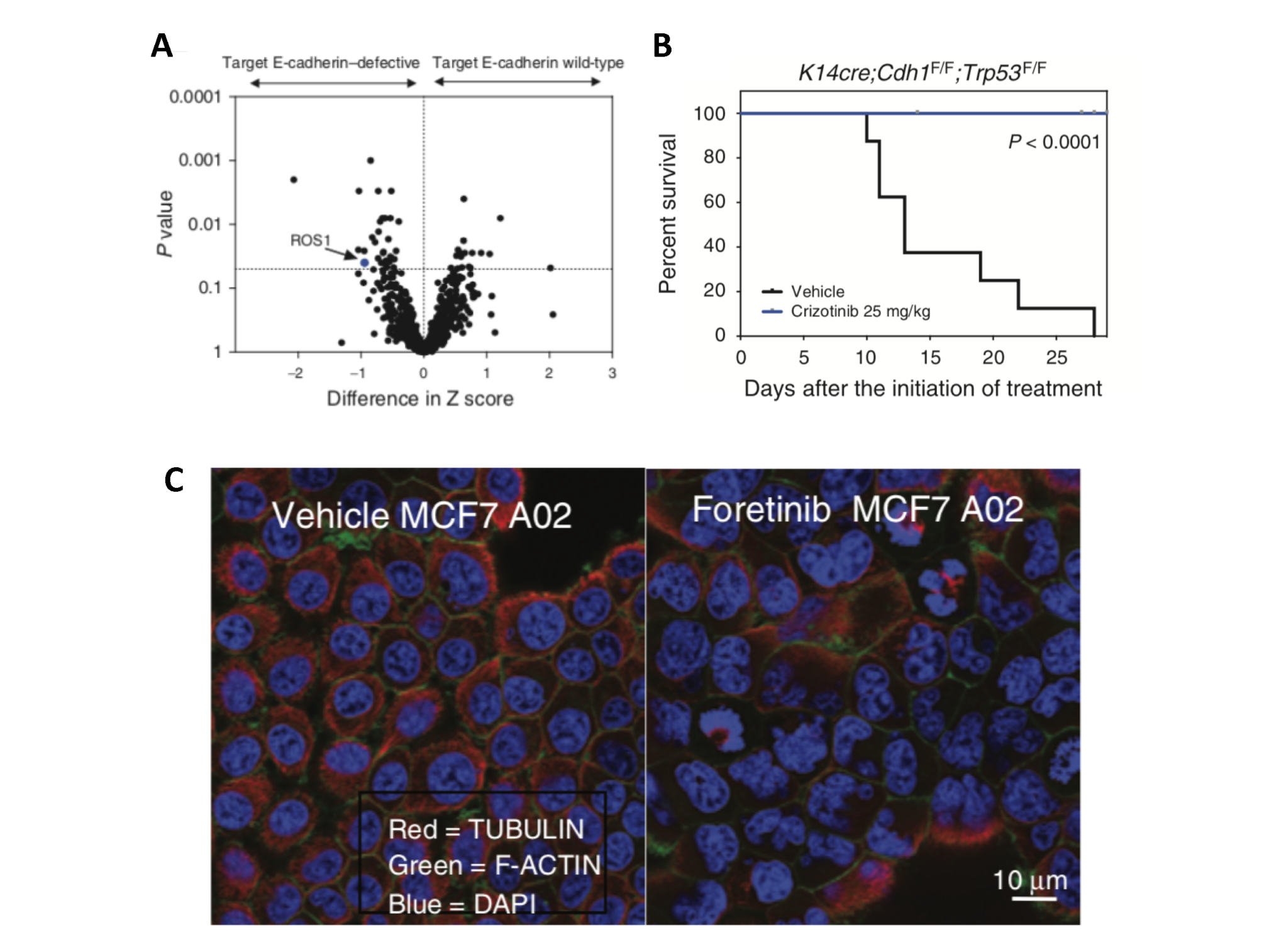
ROS1 inhibitors target E-cadherin defective cancer. A) Identification of ROS1 as a genetic dependency in E-cadherin defective tumour cell lines using an siRNA screen. B) Improved survival in a mouse model of E-cadherin (Cdh1) defective breast cancer treated with the ROS1 inhibitor crizotinib. C) Multinucleation and mitotic defects induced by the ROS1 inhibitor foretinib in E-cadherin mutant engineered MCF7 cells.
Targeting E-cadherin defects in lobular breast cancer

Building on our identification of synthetic lethality between BRCA1/BRCA2 mutations and PARP inhibitors, we are focussed on identifying synthetic lethal interactions in other breast cancer contexts.
E-cadherin defectives are particularly common in patients diagnosed with the Invasive Lobular breast cancer subtype (ILC), due to somatic CDH1 frameshift mutation, loss of heterozygosity or CDH1 promoter methylation. The fundamental aim of our research is to identify new treatment options for these breast cancer patients, utilizing synthetic lethality studies in the lab to improve their survival outcome.
We have previously identified the firstly clinically actionable synthetic lethality for breast cancers that have E-cadherin defects, through treatment with Crizotinib (Bajrami et al, Cancer Discovery 2018, Figure 3). The efficacy of this targeted treatment is currently being assessed in patients in the actively recruiting ROLo Phase 2 clinical trial (NCT03620643), led by Nicholas Turner (BCN Centre, ICR and Royal Marsden Hospital) and Alicia Okines (Royal Marsden Hospital).
One of our current aims is to understand clinical responses in the ROLo trial. Despite this recent treatment advancement, data generated from our group would suggest that a number of these patients may develop resistance to Crizotinib. We are identifying mechanisms by which E-cadherin-defective breast cancers gain resistance to Crizotinib, with the use of CRISPR screens and genomic and transcriptomic profiling of resistant models, both in vitro and in vivo.
Conversely, we would like to improve patient stratification and identify which patients are likely to respond well to Crizotinib treatment through similar Crizotinib sensitisation experiments in the lab. Additional work is also ongoing in the group to help inform the design of subsequent clinical trials, through combining Crizotinib with other drugs that are already being given to patients in different contexts.
Ongoing work in the group furthermore aims to identify new robust and highly penetrant E-cadherin defective synthetic lethal candidates, utilising CRISPRn screens and the plethora of publicly available datasets to inform the design of new breast cancer clinical trials.