
An X-ray diffraction machine
A key protein in cancer ‘self-assembles’ into structures which help drive signalling processes that fuel bowel cancer growth, a new study shows.
Scientists at The Institute of Cancer Research, London, have revealed a novel way the protein Tankyrase supports signals which help bowel cancer cells replicate out of control, by blocking mechanisms that normally prevent cell division.
They saw that regions of Tankyrase enable it to assemble into structures that can switch off so-called ‘destruction complexes’ — groups of proteins which work together to keep cell division at bay under normal conditions.
Some types of bowel cancer cells require Tankyrase to grow, so the findings may help researchers develop new drugs to target the protein.
Mutations in cancer can alter proteins and derail signalling processes involved in cell division, causing cancer to grow out of control.
The ‘destruction complex’
Bowel cancer is one of the most frequently occurring cancers worldwide, and many patients with the disease have mutations that affect proteins that are part of the ‘β-catenin destruction complex’, which regulates cell division.
In healthy bowel cells, the protein Tankyrase helps enable normal cell division by turning off the destruction complex.
In bowel cancer, mutations can stop the destruction complex from working normally, allowing proteins promoting cancer cell growth to build up unchecked. If the destruction complex could be switched back on, at least to some degree, it could be an important new way to treat the disease.
In the study funded by Cancer Research UK and the ICR, and published in the journal Molecular Cell, ICR researchers used X-ray crystallography to investigate a region of Tankyrase called the SAM domain, to understand how the protein turns off the β-catenin destruction complex.
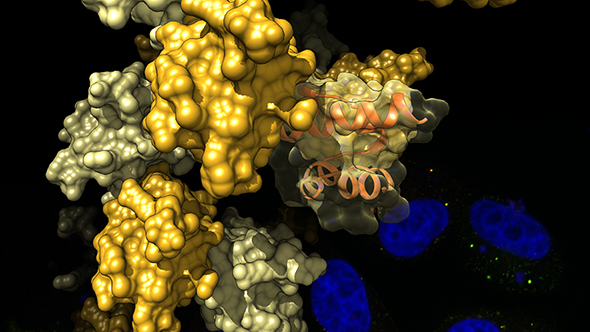
CLICK TO ENLARGE: The SAM domain of Tankyrase self-assembles into structures resembling curled pearl strings. The image shows a representation of these structures, elucidated by X-ray crystallography, with data gathered at the Diamond Light Source in Oxfordshire. The background shows cells stained for their cell nuclei (blue) and Tankyrase (red and green) (image: Dr Sebastian Guettler).
'Pearl strings'
They saw that the SAM domain enables Tankyrase molecules to assemble with each other into intriguing repetitive structures resembling curled pearl strings.
Further experiments showed these structures are essential for Tankyrase to effectively recognise and turn off destruction complexes. These domains could be exciting targets for future cancer drugs.
Study leader Dr Sebastian Guettler, Team Leader in the Divisions of Structural and Cancer Biology, said: “Our study gives important new insights into the structure of a major cancer-relevant protein called Tankyrase and its associated network of signalling molecules, providing new clues about how we might disrupt this network using future cancer drugs.
“We interrogated the structure of Tankyrase in great detail to decipher the function of potentially targetable domains in Tankyrase which allow it to turn off destruction complexes that regulate cell division. Our findings open up the tantalising possibility of restoring destruction complex activity, at least partially, by inhibiting the function of these domains. This way, we hope we will be able to target certain cancer cells in the future.”
The new study adds to a long-standing interest at The Institute of Cancer Research in studying Tankyrase as an anti-cancer drug target.