A scientist reviews a DNA stream in print. (photo: iStock.com/Claude Dagenais)
A number of recent publications in the scientific literature have reignited a longstanding debate in cancer research that focuses upon classifying genes as being either essential – i.e. absolutely required for sustaining life – or non-essential.
Rather than being an arcane question, I think that understanding the difference between these two classifications lies at the very heart of how we might better treat cancer.
Before I explain why this distinction is important, a tiny bit of background is probably required. Since the 1960s, a series of large-scale studies have mutated each of the approximately 6,000 genes in a very simple organism, yeast, to understand how each gene modifies cell behavior.
These genetic screens (see figure 1 below), which are still used today, have had the additional benefit of identifying those genes that when mutated caused cell death – the so-called ‘essential’ genes.
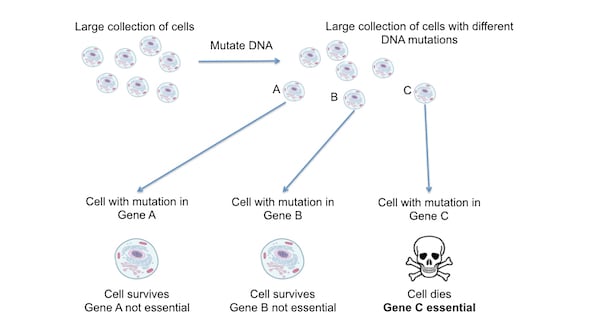
Figure 1. Genetic screens can identify essential genes. In genetic screens a large population (a collection of cells that all derived from the same original cell) are mutated using DNA technology (see later). This generates a large population of cells where each cell has a mutation in a different gene. Cells with mutations in genes A, B or C are shown for simplicity. The survival of each cell is assessed. Cells with mutations in genes A and B survive, suggesting that these genes are not essential. Contrastingly, mutation in gene C causes cell death, suggesting gene C is essential.
In yeast, somewhere in the region of 10–19% of genes are essential (the number varies slightly depending on which lab carried out the work).
When similar studies have been carried out in roundworms, fruit flies or a type of tropical fish, the zebrafish, the proportion of essential genes is also in the region of 10%.
Which genes are essential genes?
These genetic screens have also taught us something about the characteristics of essential genes. For example, regardless of the species involved, essential genes tend to control a relatively small number of fundamental activities in cells, such as converting DNA into RNA (the process known as transcription).
These core processes tend to be carried out using the same molecular machinery, whether you happen to be a fruit fly, roundworm or tropical fish.
Essential genes also tend not to accumulate DNA mutations that might alter their function, whereas non-essential genes do. Again, this seems fairly obvious – genes that control essential processes cannot afford to be altered to any great extent.
Finally, essential genes tend to be passed on through evolution from one species to the next (a concept known as gene conservation). This also makes a lot of sense, given the processes they control are essential regardless of the type of species you happen to be.
In human cells, doing effective genetic screens has until recently proven quite difficult and so this has made the classification of essential and non-essential genes somewhat complex.
Testing for essential genes with the CRISPR-Cas9 system
However, in the past two years, a technique known as CRISPR (clustered regularly interspaced short palindromic repeats)-based mutagenesis has really driven research in this area forward.
CRISPR mutagenesis uses a piece of RNA (a ‘guide’ RNA) to guide an enzyme, known as Cas9 endonuclease, to a specific part of the DNA, for example to a specific gene.
Once at the specific gene, Cas9 cuts the DNA structure (see figure 2 below). The ineffective repair of cut DNA often introduces DNA mutations into the DNA sequence, many of which render the gene dysfunctional.
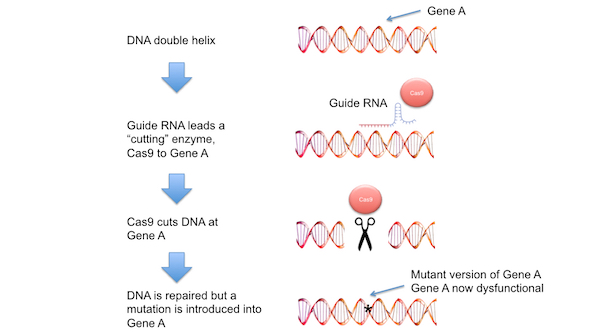
Figure 2. CRISPR-Cas9 mutagenesis. CRISPR-Cas9 technology allows specific genes in cells to be mutated. A piece of nucleic acid, known as a ‘guide RNA’ is designed to match the DNA sequence in a gene (gene A) to be mutated. This guide RNA is introduced into cells along with a bacterial enzyme known as Cas9. The guide RNA leads the Cas9 enzyme to gene A, where Cas9 cuts the DNA double helix. The cell’s normal DNA repair machinery repairs the break in the DNA double helix, but in doing so often introduces DNA mutations (incorrect or missing pieces of DNA). In this particular case, DNA mutation in gene A prevents this gene from functioning normally. Large collections of guide RNAs, each one targeting one of the 25,000 genes found in human cells, have now been developed, making genetic screens in human cells (Figure 1) a powerful experimental approach.
A number of labs have developed a series of guide RNAs that allow scientists to mutate, one-by-one, each of the 25,000 genes in the human genome. In doing so, a list of essential human genes has now been compiled (e.g. see this 2015 paper by Wang et al. or this 2016 preprint by Hart et al.).
How essential are human genes?
Looking at the list of essential genes, many of the existing rules apply and, as in yeast, essential human genes tend to be involved in a relatively small number of processes. And if you were wondering – yes, about 10% of genes are essential in human cells as they are in other species. So there is the answer – you are 10% essential and fundamentally the same (in this regard at least) as yeast. Depressing but true.
But of course it’s not that simple. When yeast cells are grown under ‘optimal’ laboratory conditions (i.e. sufficient oxygen, nutrients etc.) the 10–19% of genes that are essential are broadly the same in terms of function as those that are essential when human cells are grown under similar conditions.
However, when the environmental conditions under which cells are grown change, the same essential genes are still required plus a whole series of additional genes. In other words, some genes are essential only under certain conditions.
In fact, in studies where yeast have been exposed to more than 700 different environmental conditions, 97% of genes were found to be essential in at least one condition.
This tells us that while only a minority of genes are essential under nearly all conditions (core essential genes), most genes are essential in at least one particular context (context-dependent essential genes).
I’ve mentioned here that this ‘context’ might be the environment a cell is in (too little oxygen, too few nutrients etc.). But it’s also very likely that some genes only become essential when other genes are mutated or dysfunctional – a concept known as induced essentiality, or synthetic lethality.
Based on studies in yeast, it seems that when a single gene is mutated, somewhere in the region of 20 other genes now become essential.
How does this relate to cancer?
How do these concepts really help us in the real world? Well, one of the important challenges we face in trying to develop better treatments for cancer is in identifying ways of targeting tumour cells while leaving normal cells relatively unharmed.
The recent discovery of the core essential genes in human cells tells us that we must avoid targeting these genes with novel cancer drugs as it is very likely that this would also cause the death of normal cells as well.
That seems quite straightforward. The other piece of information we know is that tumour cells have DNA mutations in genes that are very rarely mutated in normal cells (genes known as cancer driver genes).
If we can identify and target genes that only become essential when cancer driver genes are mutated, then we stand a fighting chance of having therapies that kill tumour cells but have minimal effects on normal cells (figure 3). Genetic screens using CRISPR technology seem a fairly robust way to identify such cancer-dependent essential genes.
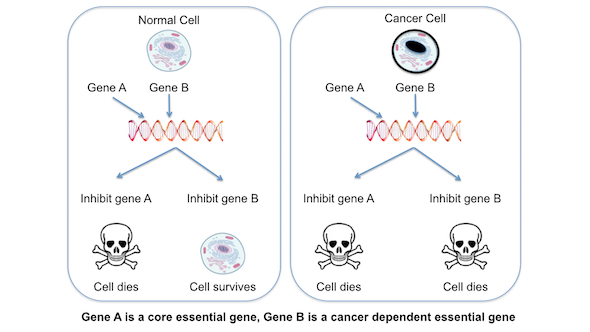
Figure 3. Cancer-dependent essential genes. In a normal cell, inhibiting gene A causes cell death, while inhibiting gene B does not. In a cancer cell, inhibiting either gene causes cell death. Gene A is essential in both cancer and normal cells and is regarded as a core essential gene, whereas gene B is only essential in cancer cells and is therefore a cancer-dependent essential gene. Gene B might only become essential in a cancer cell because of the presence of a mutation in a cancer driver gene. In principle, drugs that target gene B might selectively kill cancer cells, leaving normal cells unharmed.
Will CRISPR-Cas9 let us identify cancer-dependent essential genes?
Is doing this a reality or is this just taking a nice scientific concept too far — a kind of science fiction? A lot of our ability to detect cancer-dependent essential genes will rest on how good our genetic screening technology is, and CRISPR-Cas9 technology does look like it could deliver in this regard.
Importantly, some of the laboratories that have pioneered genetic screens using CRISPR-Cas9 are freely distributing reagents and experimental protocols that allow other labs to try this out for themselves.
This attitude should really be applauded as it is democratising the use of a novel DNA technology across the scientific community in a way we’ve not really seen before and applies a very simple ‘crowdsourcing’ logic that the more people use the technology, the faster the next iteration of CRISPR-Cas9 technology will come.
There is a note of caution though – in the past there have been a series of false dawns where the latest technique for carrying out this kind of work has promised so much but delivered so little.
A decade ago, a technology known as short hairpin RNA interference was one such new technology, which although useful, has not really lived up to its initial promise.
Will CRISPR-based techniques be the same? I hope not, and I for one am enthused by what CRISPR screens have already shown. For me, this technology looks like it might allow us to manipulate the human genome with the same ease as has been possible in yeast.
We might now be able to see whether concepts about essential, non-essential and context-dependent essential genes established in yeast operate in the cells we really want to kill — human cancer cells.
Only time (and more data) will tell.
comments powered by